Cloud-based interactive analytics for terabytes of genomic variants data
- PMID: 28961771
- PMCID: PMC5860318
- DOI: 10.1093/bioinformatics/btx468
Cloud-based interactive analytics for terabytes of genomic variants data
Abstract
Motivation: Large scale genomic sequencing is now widely used to decipher questions in diverse realms such as biological function, human diseases, evolution, ecosystems, and agriculture. With the quantity and diversity these data harbor, a robust and scalable data handling and analysis solution is desired.
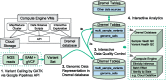
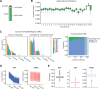
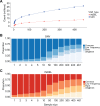
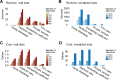
Results: We present interactive analytics using a cloud-based columnar database built on Dremel to perform information compression, comprehensive quality controls, and biological information retrieval in large volumes of genomic data. We demonstrate such Big Data computing paradigms can provide orders of magnitude faster turnaround for common genomic analyses, transforming long-running batch jobs submitted via a Linux shell into questions that can be asked from a web browser in seconds. Using this method, we assessed a study population of 475 deeply sequenced human genomes for genomic call rate, genotype and allele frequency distribution, variant density across the genome, and pharmacogenomic information.
Availability and implementation: Our analysis framework is implemented in Google Cloud Platform and BigQuery. Codes are available at https://github.com/StanfordBioinformatics/mvp_aaa_codelabs.
Contact: cuiping@stanford.edu or ptsao@stanford.edu.
Supplementary information: Supplementary data are available at Bioinformatics online.
Published by Oxford University Press 2017. This work is written by US Government employees and are in the public domain in the US.
Figures
References
-
- Abul-Husn N.S. et al. (2016) Genetic identification of familial hypercholesterolemia within a single U.S. health care system. Science (New York, N.Y.), 354. - PubMed
-
- Athanasiu L. et al. (2017) A genetic association study of CSMD1 and CSMD2 with cognitive function. Brain Behav. Immun., 61, 209–216. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

