Covert dissemination of carbapenemase-producing Klebsiella pneumoniae (KPC) in a successfully controlled outbreak: long- and short-read whole-genome sequencing demonstrate multiple genetic modes of transmission
- PMID: 28961793
- PMCID: PMC5890743
- DOI: 10.1093/jac/dkx264
Covert dissemination of carbapenemase-producing Klebsiella pneumoniae (KPC) in a successfully controlled outbreak: long- and short-read whole-genome sequencing demonstrate multiple genetic modes of transmission
Abstract
Background: Carbapenemase-producing Enterobacteriaceae (CPE), including KPC-producing Klebsiella pneumoniae (KPC-Kpn), are an increasing threat to patient safety.
Objectives: To use WGS to investigate the extent and complexity of carbapenemase gene dissemination in a controlled KPC outbreak.
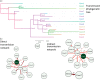
Materials and methods: Enterobacteriaceae with reduced ertapenem susceptibility recovered from rectal screening swabs/clinical samples, during a 3 month KPC outbreak (2013-14), were investigated for carbapenemase production, antimicrobial susceptibility, variable-number-tandem-repeat profile and WGS [short-read (Illumina), long-read (MinION)]. Short-read sequences were used for MLST and plasmid/Tn4401 fingerprinting, and long-read sequence assemblies for plasmid identification. Phylogenetic analysis used IQTree followed by ClonalFrameML, and outbreak transmission dynamics were inferred using SCOTTI.
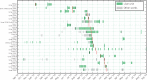
Results: Twenty patients harboured KPC-positive isolates (6 infected, 14 colonized), and 23 distinct KPC-producing Enterobacteriaceae were identified. Four distinct KPC plasmids were characterized but of 20 KPC-Kpn (from six STs), 17 isolates shared a single pKpQIL-D2 KPC plasmid. All isolates had an identical transposon (Tn4401a), except one KPC-Kpn (ST661) with a single nucleotide variant. A sporadic case of KPC-Kpn (ST491) with Tn4401a-carrying pKpQIL-D2 plasmid was identified 10 months before the outbreak. This plasmid was later seen in two other species and other KPC-Kpn (ST14,ST661) including clonal spread of KPC-Kpn (ST661) from a symptomatic case to nine ward contacts.
Conclusions: WGS of outbreak KPC isolates demonstrated blaKPC dissemination via horizontal transposition (Tn4401a), plasmid spread (pKpQIL-D2) and clonal spread (K. pneumoniae ST661). Despite rapid outbreak control, considerable dissemination of blaKPC still occurred among K. pneumoniae and other Enterobacteriaceae, emphasizing its high transmission potential and the need for enhanced control efforts.
© The Author 2017. Published by Oxford University Press on behalf of the British Society for Antimicrobial Chemotherapy.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

