A novel data-driven workflow combining literature and electronic health records to estimate comorbidities burden for a specific disease: a case study on autoimmune comorbidities in patients with celiac disease
- PMID: 28962565
- PMCID: PMC5622531
- DOI: 10.1186/s12911-017-0537-y
A novel data-driven workflow combining literature and electronic health records to estimate comorbidities burden for a specific disease: a case study on autoimmune comorbidities in patients with celiac disease
Abstract
Background: Data collected in EHRs have been widely used to identifying specific conditions; however there is still a need for methods to define comorbidities and sources to identify comorbidities burden. We propose an approach to assess comorbidities burden for a specific disease using the literature and EHR data sources in the case of autoimmune diseases in celiac disease (CD).
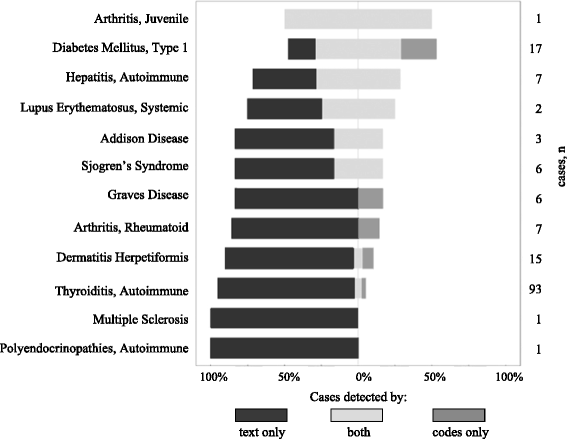
Methods: We generated a restricted set of comorbidities using the literature (via the MeSH® co-occurrence file). We extracted the 15 most co-occurring autoimmune diseases of the CD. We used mappings of the comorbidities to EHR terminologies: ICD-10 (billing codes), ATC (drugs) and UMLS (clinical reports). Finally, we extracted the concepts from the different data sources. We evaluated our approach using the correlation between prevalence estimates in our cohort and co-occurrence ranking in the literature.
Results: We retrieved the comorbidities for 741 patients with CD. 18.1% of patients had at least one of the 15 studied autoimmune disorders. Overall, 79.3% of the mapped concepts were detected only in text, 5.3% only in ICD codes and/or drugs prescriptions, and 15.4% could be found in both sources. Prevalence in our cohort were correlated with literature (Spearman's coefficient 0.789, p = 0.0005). The three most prevalent comorbidities were thyroiditis 12.6% (95% CI 10.1-14.9), type 1 diabetes 2.3% (95% CI 1.2-3.4) and dermatitis herpetiformis 2.0% (95% CI 1.0-3.0).
Conclusion: We introduced a process that leveraged the MeSH terminology to identify relevant autoimmune comorbidities of the CD and several data sources from EHRs to phenotype a large population of CD patients. We achieved prevalence estimates comparable to the literature.
Keywords: Addison disease; Antiphospholipid syndrome; Arthritis, juvenile; Arthritis, rheumatoid; Autoimmune diseases; Celiac disease; Dermatitis herpetiformis; Diabetes mellitus, type 1; Electronic health records; Graves’ disease; Hepatitis, autoimmune; Icd 10; Lupus erythematosus, systemic; Multiple sclerosis; Myasthenia gravis; Phenotype; Polyendocrinopathies, autoimmune; Prevalence study; Sjogren’s syndrome; Thyroiditis, autoimmune.
Conflict of interest statement
Ethics approval and consent to participate
This study has been approved by the institutional review board for observational study (C
Patient consent for reuse of their EHR on an opt-out basis is declared at the
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
References
-
- Benchimol EI, Guttmann A, Mack DR, Nguyen GC, Marshall JK, Gregor JC, et al. Validation of international algorithms to identify adults with inflammatory bowel disease in health administrative data from Ontario, Canada. J Clin Epidemiol. 2014;67:887–896. doi: 10.1016/j.jclinepi.2014.02.019. - DOI - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials