miR-524-5p of the primate-specific C19MC miRNA cluster targets TP53IPN1- and EMT-associated genes to regulate cellular reprogramming
- PMID: 28962647
- PMCID: PMC5622517
- DOI: 10.1186/s13287-017-0666-3
miR-524-5p of the primate-specific C19MC miRNA cluster targets TP53IPN1- and EMT-associated genes to regulate cellular reprogramming
Abstract
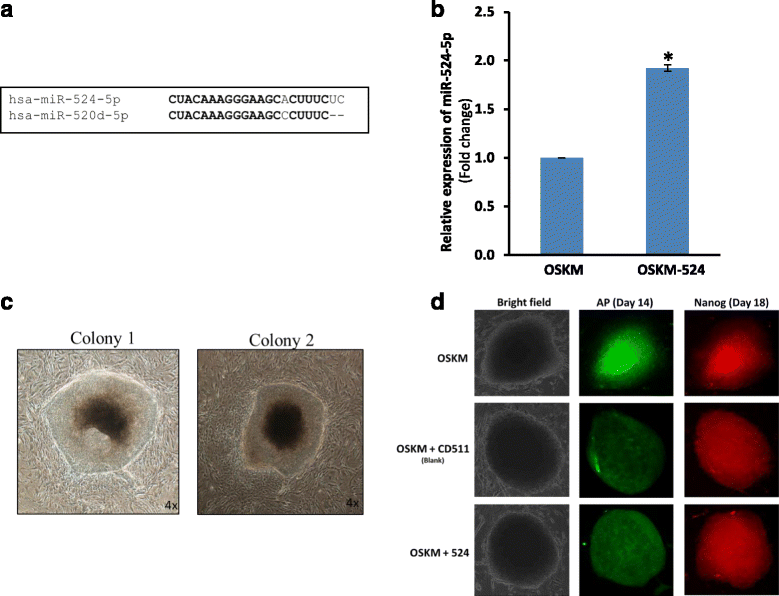
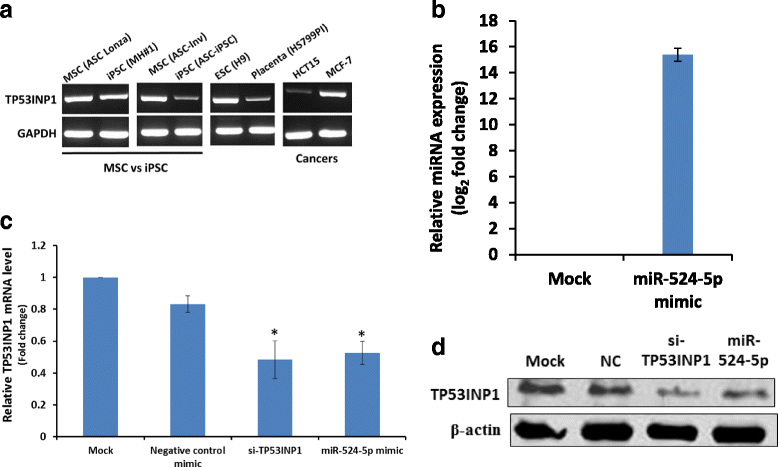
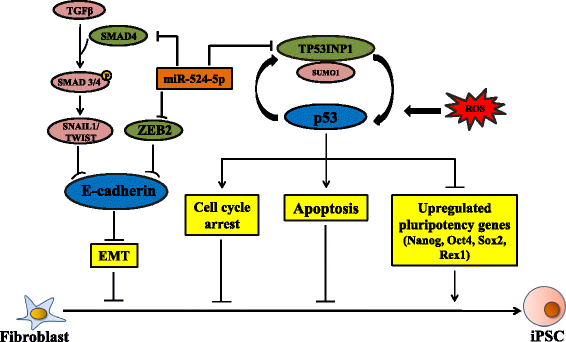
Background: Introduction of the transcription factors Oct4, Sox2, Klf4, and c-Myc (OSKM) is able to 'reprogram' somatic cells to become induced pluripotent stem cells (iPSCs). Several microRNAs (miRNAs) are known to enhance reprogramming efficiency when co-expressed with the OSKM factors. The primate-specific chromosome 19 miRNA cluster (C19MC) is essential in primate reproduction, development, and differentiation. miR-524-5p, a C19MC member, is highly homologous to the reprogramming miR-520d-5p; we also reported that miR-524-5p was expressed in iPSCs but not mesenchymal stem cells (MSCs). This study aimed to elucidate possible contributions of miR-524-5p to the reprogramming process.
Methods: A miR-524-5p precursor was introduced into human fibroblast HFF-1 in the presence of OSKM, and the relative number of embryonic stem cell (ESC)-like colonies that stained positively with alkaline phosphatase (AP) and Nanog were quantified to determine reprogramming efficiency. A miR-524-5p mimic was transfected to MSCs to investigate the effects of miR-524-5p on TP53INP1, ZEB2, and SMAD4 expression by real-time polymerase chain reaction (PCR) and Western blot. Direct gene targeting was confirmed by luciferase activity. A phylogenetic tree of TP53INP1 was constructed by the Clustal method. Contribution of miR-524-5p to cell proliferation and apoptosis was examined by cell counts, BrdU, MTT, and cell death assays, and pluripotency gene expression by real-time PCR.
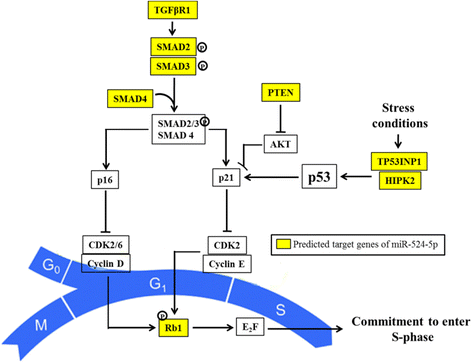
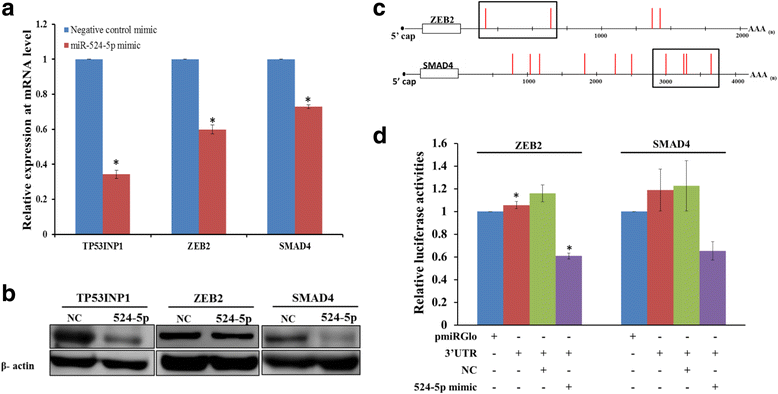
Results: Co-expressing the miR-524 precursor with OSKM resulted in a two-fold significant increase in the number of AP- and Nanog-positive ESC-like colonies, indicating a role for miR-524-5p in reprogramming. The putative target, TP53INP1, showed an inverse expression relationship with miR-524-5p; direct TP53INP1 targeting was confirmed in luciferase assays. miR-524-5p-induced TP53INP1 downregulation enhanced cell proliferation, suppressed apoptosis, and upregulated the expression of pluripotency genes, all of which are critical early events of the reprogramming process. Interestingly, the TP53INP1 gene may have co-evolved late with the primate-specific miR-524-5p. miR-524-5p also promoted mesenchymal-to-epithelial transition (MET), a required initial event of reprogramming, by directly targeting the epithelial-to-mesenchymal transition (EMT)-related genes, ZEB2 and SMAD4.
Conclusions: Via targeting TP53INP1, ZEB2, and SMAD4, miR-524-5p contributes to the early stage of inducing pluripotency by promoting cell proliferation, inhibiting apoptosis, upregulating expression of pluripotency genes, and enhancing MET. Other C19MC miRNAs may have similar reprogramming functions.
Keywords: Apoptosis; C19MC; Cell proliferation; MET; Pluripotency genes; Reprogramming efficiency; SMAD4; TP53INP1; ZEB2; miR-524-5p.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
All authors have read and given consent to the publication of the manuscript.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures


Similar articles
-
Selective activation of miRNAs of the primate-specific chromosome 19 miRNA cluster (C19MC) in cancer and stem cells and possible contribution to regulation of apoptosis.J Biomed Sci. 2017 Mar 7;24(1):20. doi: 10.1186/s12929-017-0326-z. J Biomed Sci. 2017. PMID: 28270145 Free PMC article.
-
Critical regulation of miR-200/ZEB2 pathway in Oct4/Sox2-induced mesenchymal-to-epithelial transition and induced pluripotent stem cell generation.Proc Natl Acad Sci U S A. 2013 Feb 19;110(8):2858-63. doi: 10.1073/pnas.1212769110. Epub 2013 Feb 5. Proc Natl Acad Sci U S A. 2013. PMID: 23386720 Free PMC article.
-
MiR-138-5p suppresses lung adenocarcinoma cell epithelial-mesenchymal transition, proliferation and metastasis by targeting ZEB2.Pathol Res Pract. 2019 May;215(5):861-872. doi: 10.1016/j.prp.2019.01.029. Epub 2019 Jan 28. Pathol Res Pract. 2019. PMID: 30712885
-
Learning the molecular mechanisms of the reprogramming factors: let's start from microRNAs.Mol Biosyst. 2013 Jan 27;9(1):10-7. doi: 10.1039/c2mb25088h. Epub 2012 Oct 5. Mol Biosyst. 2013. PMID: 23037570 Free PMC article. Review.
-
Regulation of the MIR155 host gene in physiological and pathological processes.Gene. 2013 Dec 10;532(1):1-12. doi: 10.1016/j.gene.2012.12.009. Epub 2012 Dec 14. Gene. 2013. PMID: 23246696 Review.
Cited by
-
Functional Targets for Epstein-Barr Virus BART MicroRNAs in B Cell Lymphomas.Cancers (Basel). 2024 Oct 19;16(20):3537. doi: 10.3390/cancers16203537. Cancers (Basel). 2024. PMID: 39456631 Free PMC article.
-
miRNAs and related genetic biomarkers according to the WHO glioma classification: From diagnosis to future therapeutic targets.Noncoding RNA Res. 2023 Oct 7;9(1):141-152. doi: 10.1016/j.ncrna.2023.10.003. eCollection 2024 Mar. Noncoding RNA Res. 2023. PMID: 38035044 Free PMC article. Review.
-
An Insight into DNA-free Reprogramming Approaches to Generate Integration-free Induced Pluripotent Stem Cells for Prospective Biomedical Applications.Stem Cell Rev Rep. 2019 Apr;15(2):286-313. doi: 10.1007/s12015-018-9861-6. Stem Cell Rev Rep. 2019. PMID: 30417242 Review.
-
The Effect of miR-106b-5p Expression in The Production of iPS-Like Cells from Mice SSCs during The Formation of Teratoma and The Three Embryonic Layers.Cell J. 2022 Aug 28;24(8):442-448. doi: 10.22074/cellj.2022.8147. Cell J. 2022. PMID: 36093803 Free PMC article.
-
The microRNA cluster C19MC confers differentiation potential into trophoblast lineages upon human pluripotent stem cells.Nat Commun. 2022 Jun 2;13(1):3071. doi: 10.1038/s41467-022-30775-w. Nat Commun. 2022. PMID: 35654791 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

