Somatic Superenhancer Duplications and Hotspot Mutations Lead to Oncogenic Activation of the KLF5 Transcription Factor
- PMID: 28963353
- PMCID: PMC5760289
- DOI: 10.1158/2159-8290.CD-17-0532
Somatic Superenhancer Duplications and Hotspot Mutations Lead to Oncogenic Activation of the KLF5 Transcription Factor
Abstract
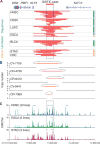
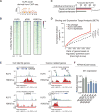
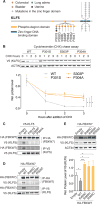
The Krüppel-like family of transcription factors plays critical roles in human development and is associated with cancer pathogenesis. Krüppel-like factor 5 gene (KLF5) has been shown to promote cancer cell proliferation and tumorigenesis and to be genomically amplified in cancer cells. We recently reported that the KLF5 gene is also subject to other types of somatic coding and noncoding genomic alterations in diverse cancer types. Here, we show that these alterations activate KLF5 by three distinct mechanisms: (i) Focal amplification of superenhancers activates KLF5 expression in squamous cell carcinomas; (ii) Missense mutations disrupt KLF5-FBXW7 interactions to increase KLF5 protein stability in colorectal cancer; (iii) Cancer type-specific hotspot mutations within a zinc-finger DNA binding domain of KLF5 change its DNA binding specificity and reshape cellular transcription. Utilizing data from CRISPR/Cas9 gene knockout screening, we reveal that cancer cells with KLF5 overexpression are dependent on KLF5 for their proliferation, suggesting KLF5 as a putative therapeutic target.Significance: Our observations, together with previous studies that identified oncogenic properties of KLF5, establish the importance of KLF5 activation in human cancers, delineate the varied genomic mechanisms underlying this occurrence, and nominate KLF5 as a putative target for therapeutic intervention in cancer. Cancer Discov; 8(1); 108-25. ©2017 AACR.This article is highlighted in the In This Issue feature, p. 1.
©2017 American Association for Cancer Research.
Conflict of interest statement
Conflict of interest: Galen F. Gao, Ashton C. Berger, Andrew D. Cherniack, and Matthew Meyerson receive research support from Bayer Pharmaceuticals. Matthew Meyerson is a consultant for and equity holder in OrigiMed. William C. Hahn receives research support from Novartis.
Figures



References
-
- Garraway LA, Widlund HR, Rubin MA, Getz G, Berger AJ, Ramaswamy S, et al. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature. 2005;436:117–22. - PubMed
-
- Visakorpi T, Hyytinen E, Koivisto P, Tanner M, Keinänen R, Palmberg C, et al. In vivo amplification of the androgen receptor gene and progression of human prostate cancer. Nat Genet. 1995;9:401–6. - PubMed
-
- Little CD, Nau MM, Carney DN, Gazdar AF, Minna JD. Amplification and expression of the c-myc oncogene in human lung cancer cell lines. Nature. 1983;306:194–6. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

