Small ocean temperature increases elicit stage-dependent changes in DNA methylation and gene expression in a fish, the European sea bass
- PMID: 28963513
- PMCID: PMC5622125
- DOI: 10.1038/s41598-017-10861-6
Small ocean temperature increases elicit stage-dependent changes in DNA methylation and gene expression in a fish, the European sea bass
Abstract
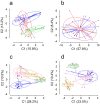
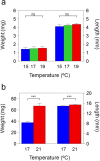
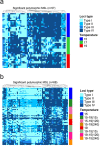
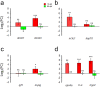
In natural fish populations, temperature increases can result in shifts in important phenotypic traits. DNA methylation is an epigenetic mechanism mediating phenotypic changes. However, whether temperature increases of the magnitude predicted by the latest global warming models can affect DNA methylation is unknown. Here, we exposed European sea bass to moderate temperature increases in different periods within the first two months of age. We show that increases of even 2 °C in larvae significantly changed global DNA methylation and the expression of ecologically-relevant genes related to DNA methylation, stress response, muscle and organ formation, while 4 °C had no effect on juveniles. Furthermore, DNA methylation changes were more marked in larvae previously acclimated to a different temperature. The expression of most genes was also affected by temperature in the larvae but not in juveniles. In conclusion, this work constitutes the first study of DNA methylation in fish showing that temperature increases of the magnitude predicted by the latest global warming models result in stage-dependent alterations in global DNA methylation and gene expression levels. This study, therefore, provides insights on the possible consequences of climate change in fish mediated by genome-wide epigenetic modifications.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
References
-
- IPCC. Climate Change 2014: Synthesis Report. Contribution of Working Groups I, II and III to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change (2014).
-
- Gilbert, S. F. & Epel, D. Ecological Developmental Biology: Integrating Epigenetics, Medicine, and Evolution. (Sinauer Associates, 2008).
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

