Initiation of ER Body Formation and Indole Glucosinolate Metabolism by the Plastidial Retrograde Signaling Metabolite, MEcPP
- PMID: 28965830
- PMCID: PMC6368977
- DOI: 10.1016/j.molp.2017.09.012
Initiation of ER Body Formation and Indole Glucosinolate Metabolism by the Plastidial Retrograde Signaling Metabolite, MEcPP
Abstract
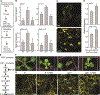
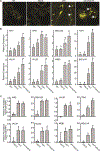
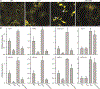
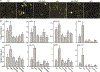
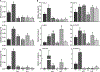
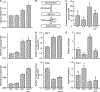
Plants have evolved tightly regulated signaling networks to respond and adapt to environmental perturbations, but the nature of the signaling hub(s) involved have remained an enigma. We have previously established that methylerythritol cyclodiphosphate (MEcPP), a precursor of plastidial isoprenoids and a stress-specific retrograde signaling metabolite, enables cellular readjustments for high-order adaptive functions. Here, we specifically show that MEcPP promotes two Brassicaceae-specific traits, namely endoplasmic reticulum (ER) body formation and induction of indole glucosinolate (IGs) metabolism selectively, via transcriptional regulation of key regulators NAI1 for ER body formation and MYB51/122 for IGs biosynthesis). The specificity of MEcPP is further confirmed by the lack of induction of wound-inducible ER body genes as well as IGs by other altered methylerythritol phosphate pathway enzymes. Genetic analyses revealed MEcPP-mediated COI1-dependent induction of these traits. Moreover, MEcPP signaling integrates the biosynthesis and hydrolysis of IGs through induction of nitrile-specifier protein1 and reduction of the suppressor, ESM1, and production of simple nitriles as the bioactive end product. The findings position the plastidial metabolite, MEcPP, as the initiation hub, transducing signals to adjust the activity of hard-wired gene circuitry to expand phytochemical diversity and alter the associated subcellular structure required for functionality of the secondary metabolites, thereby tailoring plant stress responses.
Keywords: ER body; MEP pathway; MEcPP; glucosinolates; retrograde signaling; stress.
Copyright © 2017 The Author. Published by Elsevier Inc. All rights reserved.
Figures
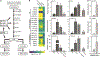

Similar articles
-
Plastidial metabolite MEcPP induces a transcriptionally centered stress-response hub via the transcription factor CAMTA3.Proc Natl Acad Sci U S A. 2016 Aug 2;113(31):8855-60. doi: 10.1073/pnas.1602582113. Epub 2016 Jul 18. Proc Natl Acad Sci U S A. 2016. PMID: 27432993 Free PMC article.
-
Plastid-produced interorgannellar stress signal MEcPP potentiates induction of the unfolded protein response in endoplasmic reticulum.Proc Natl Acad Sci U S A. 2015 May 12;112(19):6212-7. doi: 10.1073/pnas.1504828112. Epub 2015 Apr 28. Proc Natl Acad Sci U S A. 2015. PMID: 25922532 Free PMC article.
-
Retrograde signaling by the plastidial metabolite MEcPP regulates expression of nuclear stress-response genes.Cell. 2012 Jun 22;149(7):1525-35. doi: 10.1016/j.cell.2012.04.038. Cell. 2012. PMID: 22726439
-
Plastidial metabolites and retrograde signaling: A case study of MEP pathway intermediate MEcPP that orchestrates plant growth and stress responses.Plant Physiol Biochem. 2025 May;222:109747. doi: 10.1016/j.plaphy.2025.109747. Epub 2025 Mar 5. Plant Physiol Biochem. 2025. PMID: 40073740 Review.
-
Plastidial retrograde modulation of light and hormonal signaling: an odyssey.New Phytol. 2021 May;230(3):931-937. doi: 10.1111/nph.17192. Epub 2021 Feb 4. New Phytol. 2021. PMID: 33452833 Review.
Cited by
-
Uncovering the functional residues of Arabidopsis isoprenoid biosynthesis enzyme HDS.Proc Natl Acad Sci U S A. 2020 Jan 7;117(1):355-361. doi: 10.1073/pnas.1916434117. Epub 2019 Dec 26. Proc Natl Acad Sci U S A. 2020. PMID: 31879352 Free PMC article.
-
Cesium tolerance is enhanced by a chemical which binds to BETA-GLUCOSIDASE 23 in Arabidopsis thaliana.Sci Rep. 2021 Oct 26;11(1):21109. doi: 10.1038/s41598-021-00564-4. Sci Rep. 2021. PMID: 34702872 Free PMC article.
-
Orthogonal regulation of phytochrome B abundance by stress-specific plastidial retrograde signaling metabolite.Nat Commun. 2019 Jul 2;10(1):2904. doi: 10.1038/s41467-019-10867-w. Nat Commun. 2019. PMID: 31266952 Free PMC article.
-
Plastids in a Pinch: Coordinating Stress and Developmental Responses Through Retrograde Signalling.Plant Cell Environ. 2025 Sep;48(9):6897-6911. doi: 10.1111/pce.15664. Epub 2025 Jun 5. Plant Cell Environ. 2025. PMID: 40474490 Free PMC article. Review.
-
Information-incorporated gene network construction with FDR control.Bioinformatics. 2024 Mar 4;40(3):btae125. doi: 10.1093/bioinformatics/btae125. Bioinformatics. 2024. PMID: 38430463 Free PMC article.
References
-
- Ahn YO, Shimizu BI, Sakata K, Gantulga D, Zhou CH, Bevan DR, and Esen A (2010). Scopolin-hydrolyzing beta-glucosidases in roots of Arabidopsis. Plant Cell Physiol. 51:132–143, - PubMed
-
- Barth C, and Jander G (2006). Arabidopsis myrosinases TGG1 and TGG2 have redundant function in glucosinolate breakdown and insect defense. Plant J. 46:549–562. - PubMed
-
- Bednarek P (2012). Chemical warfare or modulators of defence responses - the function of secondary metabolites in plant immunity. Curr. Opin. Plant Biol. 15:407–414. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

