The Immune Signaling Adaptor LAT Contributes to the Neuroanatomical Phenotype of 16p11.2 BP2-BP3 CNVs
- PMID: 28965845
- PMCID: PMC5630231
- DOI: 10.1016/j.ajhg.2017.08.016
The Immune Signaling Adaptor LAT Contributes to the Neuroanatomical Phenotype of 16p11.2 BP2-BP3 CNVs
Abstract
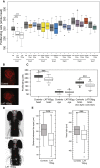
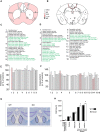
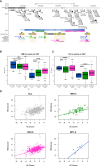
Copy-number changes in 16p11.2 contribute significantly to neuropsychiatric traits. Besides the 600 kb BP4-BP5 CNV found in 0.5%-1% of individuals with autism spectrum disorders and schizophrenia and whose rearrangement causes reciprocal defects in head size and body weight, a second distal 220 kb BP2-BP3 CNV is likewise a potent driver of neuropsychiatric, anatomical, and metabolic pathologies. These two CNVs are engaged in complex reciprocal chromatin looping, intimating a functional relationship between genes in these regions that might be relevant to pathomechanism. We assessed the drivers of the distal 16p11.2 duplication by overexpressing each of the nine encompassed genes in zebrafish. Only overexpression of LAT induced a reduction of brain proliferating cells and concomitant microcephaly. Consistently, suppression of the zebrafish ortholog induced an increase of proliferation and macrocephaly. These phenotypes were not unique to zebrafish; Lat knockout mice show brain volumetric changes. Consistent with the hypothesis that LAT dosage is relevant to the CNV pathology, we observed similar effects upon overexpression of CD247 and ZAP70, encoding members of the LAT signalosome. We also evaluated whether LAT was interacting with KCTD13, MVP, and MAPK3, major driver and modifiers of the proximal 16p11.2 600 kb BP4-BP5 syndromes, respectively. Co-injected embryos exhibited an increased microcephaly, suggesting the presence of genetic interaction. Correspondingly, carriers of 1.7 Mb BP1-BP5 rearrangements that encompass both the BP2-BP3 and BP4-BP5 loci showed more severe phenotypes. Taken together, our results suggest that LAT, besides its well-recognized function in T cell development, is a major contributor of the 16p11.2 220 kb BP2-BP3 CNV-associated neurodevelopmental phenotypes.
Keywords: 16p11.2; autism; epistasis; genome architecture; head size; obesity; zebrafish.
Copyright © 2017 American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Figures


References
-
- Zufferey F., Sherr E.H., Beckmann N.D., Hanson E., Maillard A.M., Hippolyte L., Macé A., Ferrari C., Kutalik Z., Andrieux J., Simons VIP Consortium. 16p11.2 European Consortium A 600 kb deletion syndrome at 16p11.2 leads to energy imbalance and neuropsychiatric disorders. J. Med. Genet. 2012;49:660–668. - PMC - PubMed
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

