Norovirus Cell Tropism Is Determined by Combinatorial Action of a Viral Non-structural Protein and Host Cytokine
- PMID: 28966054
- PMCID: PMC5679710
- DOI: 10.1016/j.chom.2017.08.021
Norovirus Cell Tropism Is Determined by Combinatorial Action of a Viral Non-structural Protein and Host Cytokine
Abstract
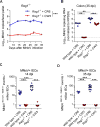
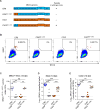
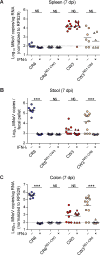
Cellular tropism during persistent viral infection is commonly conferred by the interaction of a viral surface protein with a host receptor complex. Norovirus, the leading global cause of gastroenteritis, can be persistently shed during infection, but its in vivo cellular tropism and tropism determinants remain unidentified. Using murine norovirus (MNoV), we determine that a small number of intestinal epithelial cells (IECs) serve as the reservoir for fecal shedding and persistence. The viral non-structural protein NS1, rather than a viral surface protein, determines IEC tropism. Expression of NS1 from a persistent MNoV strain is sufficient for an acute MNoV strain to target IECs and persist. In addition, interferon-lambda (IFN-λ) is a key host determinant blocking MNoV infection in IECs. The inability of acute MNoV to shed and persist is rescued in Ifnlr1-/- mice, suggesting that NS1 evades IFN-λ-mediated antiviral immunity. Thus, NS1 and IFN-λ interactions govern IEC tropism and persistence of MNoV.
Keywords: interferon-lambda; norovirus; persistence; reservoir; tropism.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures


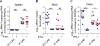
Comment in
-
IFN-Lambda: The Key to Norovirus's Secret Hideaway.Cell Host Microbe. 2017 Oct 11;22(4):427-429. doi: 10.1016/j.chom.2017.09.010. Cell Host Microbe. 2017. PMID: 29024636
Similar articles
-
CD300lf Conditional Knockout Mouse Reveals Strain-Specific Cellular Tropism of Murine Norovirus.J Virol. 2021 Jan 13;95(3):e01652-20. doi: 10.1128/JVI.01652-20. Print 2021 Jan 13. J Virol. 2021. PMID: 33177207 Free PMC article.
-
Expression of Ifnlr1 on Intestinal Epithelial Cells Is Critical to the Antiviral Effects of Interferon Lambda against Norovirus and Reovirus.J Virol. 2017 Mar 13;91(7):e02079-16. doi: 10.1128/JVI.02079-16. Print 2017 Apr 1. J Virol. 2017. PMID: 28077655 Free PMC article.
-
A Secreted Viral Nonstructural Protein Determines Intestinal Norovirus Pathogenesis.Cell Host Microbe. 2019 Jun 12;25(6):845-857.e5. doi: 10.1016/j.chom.2019.04.005. Epub 2019 May 23. Cell Host Microbe. 2019. PMID: 31130511 Free PMC article.
-
Recent advances in understanding norovirus pathogenesis.J Med Virol. 2016 Nov;88(11):1837-43. doi: 10.1002/jmv.24559. Epub 2016 May 5. J Med Virol. 2016. PMID: 27110852 Free PMC article. Review.
-
Norovirus Attachment and Entry.Viruses. 2019 May 30;11(6):495. doi: 10.3390/v11060495. Viruses. 2019. PMID: 31151248 Free PMC article. Review.
Cited by
-
Murine norovirus virulence factor 1 (VF1) protein contributes to viral fitness during persistent infection.J Gen Virol. 2021 Sep;102(9):001651. doi: 10.1099/jgv.0.001651. J Gen Virol. 2021. PMID: 34491891 Free PMC article.
-
Intestinal tuft cell immune privilege enables norovirus persistence.Sci Immunol. 2024 Mar 22;9(93):eadi7038. doi: 10.1126/sciimmunol.adi7038. Epub 2024 Mar 22. Sci Immunol. 2024. PMID: 38517952 Free PMC article.
-
Ginsenoside Rg3 enriches SCFA-producing commensal bacteria to confer protection against enteric viral infection via the cGAS-STING-type I IFN axis.ISME J. 2023 Dec;17(12):2426-2440. doi: 10.1038/s41396-023-01541-7. Epub 2023 Nov 10. ISME J. 2023. PMID: 37950067 Free PMC article.
-
Caspase-mediated cleavage of murine norovirus NS1/2 potentiates apoptosis and is required for persistent infection of intestinal epithelial cells.PLoS Pathog. 2019 Jul 22;15(7):e1007940. doi: 10.1371/journal.ppat.1007940. eCollection 2019 Jul. PLoS Pathog. 2019. PMID: 31329638 Free PMC article.
-
The intestinal regionalization of acute norovirus infection is regulated by the microbiota via bile acid-mediated priming of type III interferon.Nat Microbiol. 2020 Jan;5(1):84-92. doi: 10.1038/s41564-019-0602-7. Epub 2019 Nov 25. Nat Microbiol. 2020. PMID: 31768030 Free PMC article.
References
-
- Ank N, Iversen MB, Bartholdy C, Staeheli P, Hartmann R, Jensen UB, Dagnaes-Hansen F, Thomsen AR, Chen Z, Haugen H, et al. An important role for type III interferon (IFN-lambda/IL-28) in TLR-induced antiviral activity. Journal of immunology. 2008;180:2474–2485. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

