Neurochemical Heterogeneity Among Lateral Hypothalamic Hypocretin/Orexin and Melanin-Concentrating Hormone Neurons Identified Through Single-Cell Gene Expression Analysis
- PMID: 28966976
- PMCID: PMC5617207
- DOI: 10.1523/ENEURO.0013-17.2017
Neurochemical Heterogeneity Among Lateral Hypothalamic Hypocretin/Orexin and Melanin-Concentrating Hormone Neurons Identified Through Single-Cell Gene Expression Analysis
Abstract
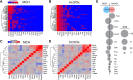
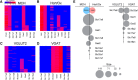
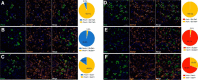
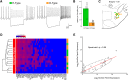
The lateral hypothalamic area (LHA) lies at the intersection of multiple neural and humoral systems and orchestrates fundamental aspects of behavior. Two neuronal cell types found in the LHA are defined by their expression of hypocretin/orexin (Hcrt/Ox) and melanin-concentrating hormone (MCH) and are both important regulators of arousal, feeding, and metabolism. Conflicting evidence suggests that these cell populations have a more complex signaling repertoire than previously appreciated, particularly in regard to their coexpression of other neuropeptides and the machinery for the synthesis and release of GABA and glutamate. Here, we undertook a single-cell expression profiling approach to decipher the neurochemical phenotype, and heterogeneity therein, of Hcrt/Ox and MCH neurons. In transgenic mouse lines, we used single-cell quantitative polymerase chain reaction (qPCR) to quantify the expression of 48 key genes, which include neuropeptides, fast neurotransmitter components, and other key markers, which revealed unexpected neurochemical diversity. We found that single MCH and Hcrt/Ox neurons express transcripts for multiple neuropeptides and markers of both excitatory and inhibitory fast neurotransmission. Virtually all MCH and approximately half of the Hcrt/Ox neurons sampled express both the machinery for glutamate release and GABA synthesis in the absence of a vesicular GABA release pathway. Furthermore, we found that this profile is characteristic of a subpopulation of LHA glutamatergic neurons but contrasts with a broad population of LHA GABAergic neurons. Identifying the neurochemical diversity of Hcrt/Ox and MCH neurons will further our understanding of how these populations modulate postsynaptic excitability through multiple signaling mechanisms and coordinate diverse behavioral outputs.
Keywords: cotransmission; hypocretin/orexin; lateral hypothalamic area; melanin-concentrating hormone; neuropeptide; neurotransmitter.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
