Evaluation of the innate immune responses to influenza and live-attenuated influenza vaccine infection in primary differentiated human nasal epithelial cells
- PMID: 28967519
- PMCID: PMC5647870
- DOI: 10.1016/j.vaccine.2017.09.058
Evaluation of the innate immune responses to influenza and live-attenuated influenza vaccine infection in primary differentiated human nasal epithelial cells
Abstract
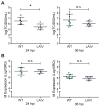
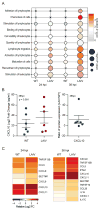
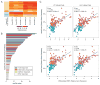
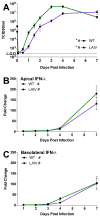
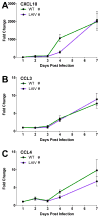
The host innate immune response to influenza virus is a key determinant of pathogenic outcomes and long-term protective immune responses against subsequent exposures. Here, we present a direct contrast of the host responses in primary differentiated human nasal epithelial cell (hNEC) cultures following infection with either a seasonal H3N2 influenza virus (WT) or the antigenically-matched live-attenuated vaccine (LAIV) strain. Comparison of the transcriptional profiles obtained 24 and 36h post-infection showed that the magnitude of gene expression was greater in LAIV infected relative to that observed in WT infected hNEC cultures. Functional enrichment analysis revealed that the antiviral and inflammatory responses were largely driven by type III IFN induction in both WT and LAIV infected cells. However, the enrichment of biological pathways involved in the recruitment of mononuclear leukocytes, antigen-presenting cells, and T lymphocytes was uniquely observed in LAIV infected cells. These observations were reflective of the host innate immune responses observed in individuals acutely infected with influenza viruses. These findings indicate that cell-intrinsic type III IFN-mediated innate immune responses in the nasal epithelium are not only crucial for viral clearance and attenuation, but may also play an important role in the induction of protective immune responses with live-attenuated vaccines.
Keywords: Epithelial cells; Host response; Inflammation; Influenza virus; Interferon; Live-attenuated vaccine.
Copyright © 2017 Elsevier Ltd. All rights reserved.
Figures
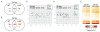

Similar articles
-
Live attenuated influenza vaccine strains elicit a greater innate immune response than antigenically-matched seasonal influenza viruses during infection of human nasal epithelial cell cultures.Vaccine. 2014 Mar 26;32(15):1761-7. doi: 10.1016/j.vaccine.2013.12.069. Epub 2014 Jan 30. Vaccine. 2014. PMID: 24486351 Free PMC article.
-
Restricted replication of the live attenuated influenza A virus vaccine during infection of primary differentiated human nasal epithelial cells.Vaccine. 2015 Aug 26;33(36):4495-504. doi: 10.1016/j.vaccine.2015.07.023. Epub 2015 Jul 18. Vaccine. 2015. PMID: 26196325 Free PMC article.
-
Electronic-Cigarette Use Alters Nasal Mucosal Immune Response to Live-attenuated Influenza Virus. A Clinical Trial.Am J Respir Cell Mol Biol. 2021 Jan;64(1):126-137. doi: 10.1165/rcmb.2020-0164OC. Am J Respir Cell Mol Biol. 2021. PMID: 33095645 Free PMC article.
-
Temperature Sensitive Mutations in Influenza A Viral Ribonucleoprotein Complex Responsible for the Attenuation of the Live Attenuated Influenza Vaccine.Viruses. 2018 Oct 15;10(10):560. doi: 10.3390/v10100560. Viruses. 2018. PMID: 30326610 Free PMC article. Review.
-
Live attenuated vaccines for pandemic influenza.Curr Top Microbiol Immunol. 2009;333:109-32. doi: 10.1007/978-3-540-92165-3_5. Curr Top Microbiol Immunol. 2009. PMID: 19768402 Review.
Cited by
-
2019-20 H1N1 clade A5a.1 viruses have better in vitro replication compared with the co-circulating A5a.2 clade.bioRxiv [Preprint]. 2023 Feb 26:2023.02.26.530085. doi: 10.1101/2023.02.26.530085. bioRxiv. 2023. Update in: Sci Rep. 2023 Jun 23;13(1):10223. doi: 10.1038/s41598-023-37122-z. PMID: 36865250 Free PMC article. Updated. Preprint.
-
Wood Smoke Exposure Alters Human Inflammatory Responses to Viral Infection in a Sex-Specific Manner. A Randomized, Placebo-controlled Study.Am J Respir Crit Care Med. 2019 Apr 15;199(8):996-1007. doi: 10.1164/rccm.201807-1287OC. Am J Respir Crit Care Med. 2019. PMID: 30360637 Free PMC article. Clinical Trial.
-
Oncostatin M Contributes to Airway Epithelial Cell Dysfunction in Chronic Rhinosinusitis with Nasal Polyps.Int J Mol Sci. 2023 Mar 23;24(7):6094. doi: 10.3390/ijms24076094. Int J Mol Sci. 2023. PMID: 37047067 Free PMC article.
-
Sequential Infection with Influenza A Virus Followed by Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Leads to More Severe Disease and Encephalitis in a Mouse Model of COVID-19.Viruses. 2024 May 28;16(6):863. doi: 10.3390/v16060863. Viruses. 2024. PMID: 38932155 Free PMC article.
-
Associated virus-bacterial vaccine based on seasonal LAIV and S. pneumoniae chimeric peptide provide protection against post-influenza pneumococcal infection in mouse model.Virulence. 2022 Dec;13(1):558-568. doi: 10.1080/21505594.2022.2049496. Virulence. 2022. PMID: 35266442 Free PMC article.
References
-
- Ank N, Iversen MB, Bartholdy C, Staeheli P, Hartmann R, Jensen UB, Dagnaes-Hansen F, Thomsen AR, Chen Z, Haugen H, Klucher K, Paludan SR. An important role for type III interferon (IFN-lambda/IL-28) in TLR-induced antiviral activity. J Immunol. 2008;180:2474–2485. - PubMed
-
- Benjamini Y, Hochberg Y. Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. J Roy Stat Soc B Met. 1995;57:289–300.
-
- Chan W, Zhou H, Kemble G, Jin H. The cold adapted and temperature sensitive influenza A/Ann Arbor/6/60 virus, the master donor virus for live attenuated influenza vaccines, has multiple defects in replication at the restrictive temperature. Virology. 2008;380:304–311. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

