Association score testing for rare variants and binary traits in family data with shared controls
- PMID: 28968627
- PMCID: PMC6357552
- DOI: 10.1093/bib/bbx107
Association score testing for rare variants and binary traits in family data with shared controls
Abstract
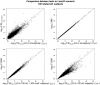
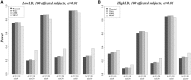
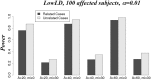
Genome-wide association studies have been an important approach used to localize trait loci, with primary focus on common variants. The multiple rare variant-common disease hypothesis may explain the missing heritability remaining after accounting for identified common variants. Advances of sequencing technologies with their decreasing costs, coupled with methodological advances in the context of association studies in large samples, now make the study of rare variants at a genome-wide scale feasible. The resurgence of family-based association designs because of their advantage in studying rare variants has also stimulated more methods development, mainly based on linear mixed models (LMMs). Other tests such as score tests can have advantages over the LMMs, but to date have mainly been proposed for single-marker association tests. In this article, we extend several score tests (χcorrected2, WQLS, and SKAT) to the multiple variant association framework. We evaluate and compare their statistical performances relative with the LMM. Moreover, we show that three tests can be cast as the difference between marker allele frequencies (AFs) estimated in each of the group of affected and unaffected subjects. We show that these tests are flexible, as they can be based on related, unrelated or both related and unrelated subjects. They also make feasible an increasingly common design that only sequences a subset of affected subjects (related or unrelated) and uses for comparison publicly available AFs estimated in a group of healthy subjects. Finally, we show the great impact of linkage disequilibrium on the performance of all these tests.
Figures
Similar articles
-
Combining family- and population-based imputation data for association analysis of rare and common variants in large pedigrees.Genet Epidemiol. 2014 Nov;38(7):579-90. doi: 10.1002/gepi.21844. Epub 2014 Aug 1. Genet Epidemiol. 2014. PMID: 25132070 Free PMC article.
-
Rare variant association test with multiple phenotypes.Genet Epidemiol. 2017 Apr;41(3):198-209. doi: 10.1002/gepi.22021. Epub 2016 Dec 31. Genet Epidemiol. 2017. PMID: 28039885 Free PMC article.
-
Joint association analysis of a binary and a quantitative trait in family samples.Eur J Hum Genet. 2016 Jan;25(1):130-136. doi: 10.1038/ejhg.2016.134. Epub 2016 Oct 26. Eur J Hum Genet. 2016. PMID: 27782109 Free PMC article.
-
Association Tests for Rare Variants.Annu Rev Genomics Hum Genet. 2016 Aug 31;17:117-30. doi: 10.1146/annurev-genom-083115-022609. Epub 2016 Apr 21. Annu Rev Genomics Hum Genet. 2016. PMID: 27147090 Review.
-
Identifying rare variants associated with complex traits via sequencing.Curr Protoc Hum Genet. 2013 Jul;Chapter 1:Unit 1.26. doi: 10.1002/0471142905.hg0126s78. Curr Protoc Hum Genet. 2013. PMID: 23853079 Free PMC article. Review.
Cited by
-
A novel rare variants association test for binary traits in family-based designs via copulas.Stat Methods Med Res. 2023 Nov;32(11):2096-2122. doi: 10.1177/09622802231197977. Epub 2023 Oct 13. Stat Methods Med Res. 2023. PMID: 37832140 Free PMC article.
-
An Efficient Bayesian Method for Estimating the Degree of the Skewness of X Chromosome Inactivation Based on the Mixture of General Pedigrees and Unrelated Females.Biomolecules. 2023 Mar 16;13(3):543. doi: 10.3390/biom13030543. Biomolecules. 2023. PMID: 36979477 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

