VirGenA: a reference-based assembler for variable viral genomes
- PMID: 28968771
- PMCID: PMC6488938
- DOI: 10.1093/bib/bbx079
VirGenA: a reference-based assembler for variable viral genomes
Abstract
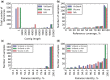
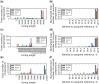
Characterization of the within-host genetic diversity of viral pathogens is required for selection of effective treatment of some important viral infections, e.g. HIV, HBV and HCV. Despite the technical ability of detection, there are conflicting data regarding the clinical significance of low-frequency variants, partially because of the difficulty of their distinguishing from experimental artifacts. The issue of cross-contamination is relevant for all highly sensitive techniques, including deep sequencing: even trace contamination leads to a significant increase of false positives in identified SNVs. Determination of infections by multiple genotypes of some viruses, the incidence of which can be considerable, especially in risk groups, is also clinically significant in some cases. We developed a new viral reference-guided assembler, VirGenA, that can separate mixtures of strains of different intraspecies genetic groups (genotypes, subtypes, clades, etc.) and assemble a separate consensus sequence for each group in a mixture. It produced long assemblies for mixture components of extremely low frequencies (<1%) allowing detection of cross-contamination of samples by divergent genotypes. We tested VirGenA on both clinical and simulated data. On both types of data, VirGenA shows better or similar results than the existing de novo assemblers. Cross-platform implementation (including source code) is freely available at https://github.com/gFedonin/VirGenA/releases.
Figures

Similar articles
-
Viral deep sequencing needs an adaptive approach: IRMA, the iterative refinement meta-assembler.BMC Genomics. 2016 Sep 5;17(1):708. doi: 10.1186/s12864-016-3030-6. BMC Genomics. 2016. PMID: 27595578 Free PMC article.
-
QQ-SNV: single nucleotide variant detection at low frequency by comparing the quality quantiles.BMC Bioinformatics. 2015 Nov 10;16:379. doi: 10.1186/s12859-015-0812-9. BMC Bioinformatics. 2015. PMID: 26554718 Free PMC article.
-
IVA: accurate de novo assembly of RNA virus genomes.Bioinformatics. 2015 Jul 15;31(14):2374-6. doi: 10.1093/bioinformatics/btv120. Epub 2015 Feb 28. Bioinformatics. 2015. PMID: 25725497 Free PMC article.
-
Streamlined Subpopulation, Subtype, and Recombination Analysis of HIV-1 Half-Genome Sequences Generated by High-Throughput Sequencing.mSphere. 2020 Oct 14;5(5):e00551-20. doi: 10.1128/mSphere.00551-20. mSphere. 2020. PMID: 33055255 Free PMC article.
-
Clinically relevant sequence-based genotyping of HBV, HCV, CMV, and HIV.J Clin Virol. 2001 Aug;22(1):11-29. doi: 10.1016/s1386-6532(01)00156-1. J Clin Virol. 2001. PMID: 11418349 Review.
Cited by
-
Haploflow: strain-resolved de novo assembly of viral genomes.Genome Biol. 2021 Jul 19;22(1):212. doi: 10.1186/s13059-021-02426-8. Genome Biol. 2021. PMID: 34281604 Free PMC article. No abstract available.
-
Haploflow: Strain-resolved de novo assembly of viral genomes.bioRxiv [Preprint]. 2021 Jan 26:2021.01.25.428049. doi: 10.1101/2021.01.25.428049. bioRxiv. 2021. Update in: Genome Biol. 2021 Jul 19;22(1):212. doi: 10.1186/s13059-021-02426-8. PMID: 33532769 Free PMC article. Updated. Preprint.
-
New approaches for metagenome assembly with short reads.Brief Bioinform. 2020 Mar 23;21(2):584-594. doi: 10.1093/bib/bbz020. Brief Bioinform. 2020. PMID: 30815668 Free PMC article. Review.
-
Molecular Epidemiology of HIV-1 Subtype G in the Russian Federation.Viruses. 2019 Apr 16;11(4):348. doi: 10.3390/v11040348. Viruses. 2019. PMID: 30995717 Free PMC article.
-
VIGA: a one-stop tool for eukaryotic virus identification and genome assembly from next-generation-sequencing data.Brief Bioinform. 2023 Nov 22;25(1):bbad444. doi: 10.1093/bib/bbad444. Brief Bioinform. 2023. PMID: 38048079 Free PMC article.
References
-
- Department of Health and Human Services. Panel on antiretroviral guidelines for adults and adolescents Guidelines for the Use of Antiretroviral Agents in HIV-1-infected Adults and Adolescents. Department of Health and Human Services; http://www.aidsinfo.nih.gov/ContentFiles/AdultandAdolescentGL.pdf. 2016.
-
- AASLD/IDSA HCV Guidance Panel. Hepatitis C guidance: AASLD-IDSA recommendations for testing, managing, and treating adults infected with hepatitis C virus: HEPATITIS C VIRUS GUIDANCE PANEL. Hepatology 2015;62:932–54. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

