Epigenetic regulation of gene expression in cancer: techniques, resources and analysis
- PMID: 28968850
- PMCID: PMC5860551
- DOI: 10.1093/bfgp/elx018
Epigenetic regulation of gene expression in cancer: techniques, resources and analysis
Abstract
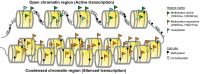
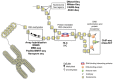
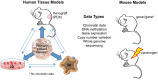
Cancer is a complex disease, driven by aberrant activity in numerous signaling pathways in even individual malignant cells. Epigenetic changes are critical mediators of these functional changes that drive and maintain the malignant phenotype. Changes in DNA methylation, histone acetylation and methylation, noncoding RNAs, posttranslational modifications are all epigenetic drivers in cancer, independent of changes in the DNA sequence. These epigenetic alterations were once thought to be crucial only for the malignant phenotype maintenance. Now, epigenetic alterations are also recognized as critical for disrupting essential pathways that protect the cells from uncontrolled growth, longer survival and establishment in distant sites from the original tissue. In this review, we focus on DNA methylation and chromatin structure in cancer. The precise functional role of these alterations is an area of active research using emerging high-throughput approaches and bioinformatics analysis tools. Therefore, this review also describes these high-throughput measurement technologies, public domain databases for high-throughput epigenetic data in tumors and model systems and bioinformatics algorithms for their analysis. Advances in bioinformatics data that combine these epigenetic data with genomics data are essential to infer the function of specific epigenetic alterations in cancer. These integrative algorithms are also a focus of this review. Future studies using these emerging technologies will elucidate how alterations in the cancer epigenome cooperate with genetic aberrations during tumor initiation and progression. This deeper understanding is essential to future studies with epigenetics biomarkers and precision medicine using emerging epigenetic therapies.
Keywords: bioinformatics; cancer; chromatin; data integration; epigenetics; methylation.
© The Author 2017. Published by Oxford University Press.
Figures

Similar articles
-
Investigation of epigenetics in kidney cell biology.Methods Cell Biol. 2019;153:255-278. doi: 10.1016/bs.mcb.2019.04.015. Epub 2019 Jun 13. Methods Cell Biol. 2019. PMID: 31395383 Free PMC article. Review.
-
Molecular and Cellular Changes During Cancer Progression Resulting From Genetic and Epigenetic Alterations.Prog Mol Biol Transl Sci. 2016;144:3-47. doi: 10.1016/bs.pmbts.2016.09.001. Epub 2016 Oct 7. Prog Mol Biol Transl Sci. 2016. PMID: 27865461 Review.
-
Epigenetic Determinants of Cancer.Cold Spring Harb Perspect Biol. 2016 Sep 1;8(9):a019505. doi: 10.1101/cshperspect.a019505. Cold Spring Harb Perspect Biol. 2016. PMID: 27194046 Free PMC article. Review.
-
Epigenetic modifications in cancer.Clin Genet. 2012 Apr;81(4):303-11. doi: 10.1111/j.1399-0004.2011.01809.x. Epub 2011 Dec 8. Clin Genet. 2012. PMID: 22082348 Free PMC article. Review.
-
The cancer epigenome--components and functional correlates.Genes Dev. 2006 Dec 1;20(23):3215-31. doi: 10.1101/gad.1464906. Genes Dev. 2006. PMID: 17158741 Review.
Cited by
-
FOXC1 Downregulates Nanog Expression by Recruiting HDAC2 to Its Promoter in F9 Cells Treated by Retinoic Acid.Int J Mol Sci. 2021 Feb 24;22(5):2255. doi: 10.3390/ijms22052255. Int J Mol Sci. 2021. PMID: 33668324 Free PMC article.
-
PDIA3 correlates with clinical malignant features and immune signature in human gliomas.Aging (Albany NY). 2020 Aug 29;12(15):15392-15413. doi: 10.18632/aging.103601. Aging (Albany NY). 2020. PMID: 32687065 Free PMC article.
-
Evidence supporting the oncogenic role of BAZ1B in colorectal cancer.Am J Cancer Res. 2022 Oct 15;12(10):4751-4763. eCollection 2022. Am J Cancer Res. 2022. PMID: 36381331 Free PMC article.
-
Increased Expression of Lysine-Specific Demethylase 5B (KDM5B) Promotes Tumor Cell Growth in Hep3B Cells and is an Independent Prognostic Factor in Patients with Hepatocellular Carcinoma.Med Sci Monit. 2018 Oct 24;24:7586-7594. doi: 10.12659/MSM.910844. Med Sci Monit. 2018. PMID: 30353907 Free PMC article.
-
Biomarker-guided heterogeneity analysis of genetic regulations via multivariate sparse fusion.Stat Med. 2021 Jul 30;40(17):3915-3936. doi: 10.1002/sim.9006. Epub 2021 Apr 27. Stat Med. 2021. PMID: 33906263 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

