Human evolution: the non-coding revolution
- PMID: 28969617
- PMCID: PMC5625771
- DOI: 10.1186/s12915-017-0428-9
Human evolution: the non-coding revolution
Abstract
What made us human? Gene expression changes clearly played a significant part in human evolution, but pinpointing the causal regulatory mutations is hard. Comparative genomics enabled the identification of human accelerated regions (HARs) and other human-specific genome sequences. The major challenge in the past decade has been to link diverged sequences to uniquely human biology. This review discusses approaches to this problem, progress made at the molecular level, and prospects for moving towards genetic causes for uniquely human biology.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

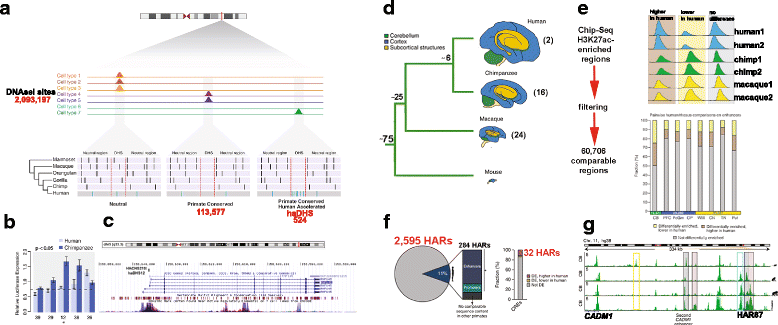
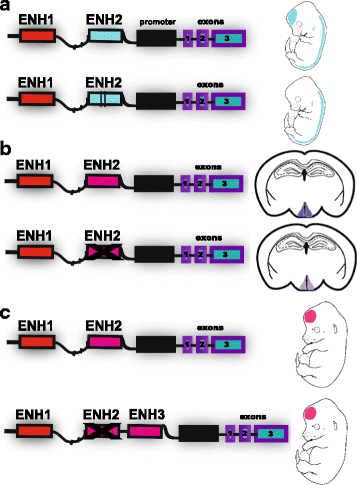
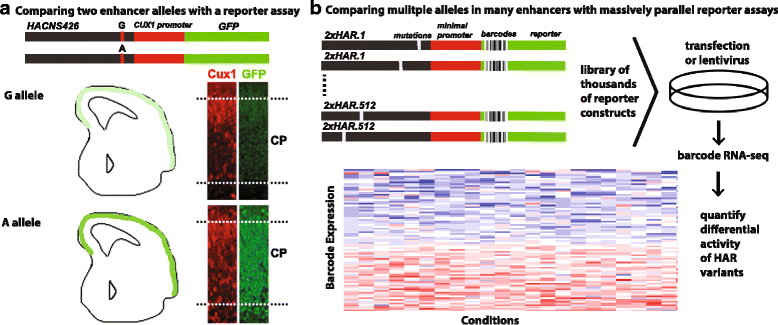
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

