Viral metagenomics of aphids present in bean and maize plots on mixed-use farms in Kenya reveals the presence of three dicistroviruses including a novel Big Sioux River virus-like dicistrovirus
- PMID: 28969654
- PMCID: PMC5625602
- DOI: 10.1186/s12985-017-0854-x
Viral metagenomics of aphids present in bean and maize plots on mixed-use farms in Kenya reveals the presence of three dicistroviruses including a novel Big Sioux River virus-like dicistrovirus
Abstract
Background: Aphids are major vectors of plant viruses. Common bean (Phaseolus vulgaris L.) and maize (Zea mays L.) are important crops that are vulnerable to aphid herbivory and aphid-transmitted viruses. In East and Central Africa, common bean is frequently intercropped by smallholder farmers to provide fixed nitrogen for cultivation of starch crops such as maize. We used a PCR-based technique to identify aphids prevalent in smallholder bean farms and next generation sequencing shotgun metagenomics to examine the diversity of viruses present in aphids and in maize leaf samples. Samples were collected from farms in Kenya in a range of agro-ecological zones.
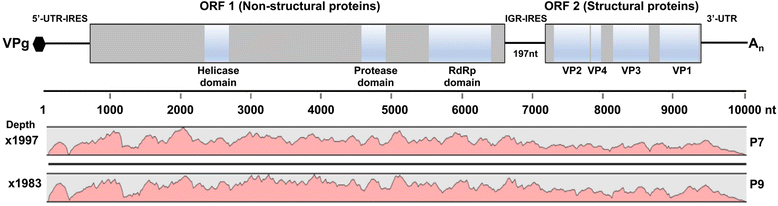
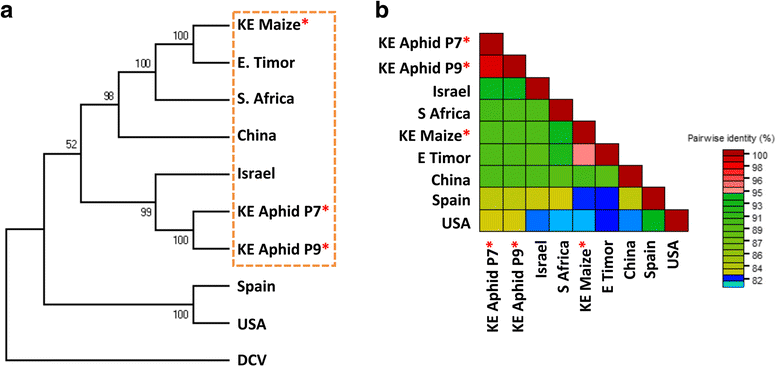
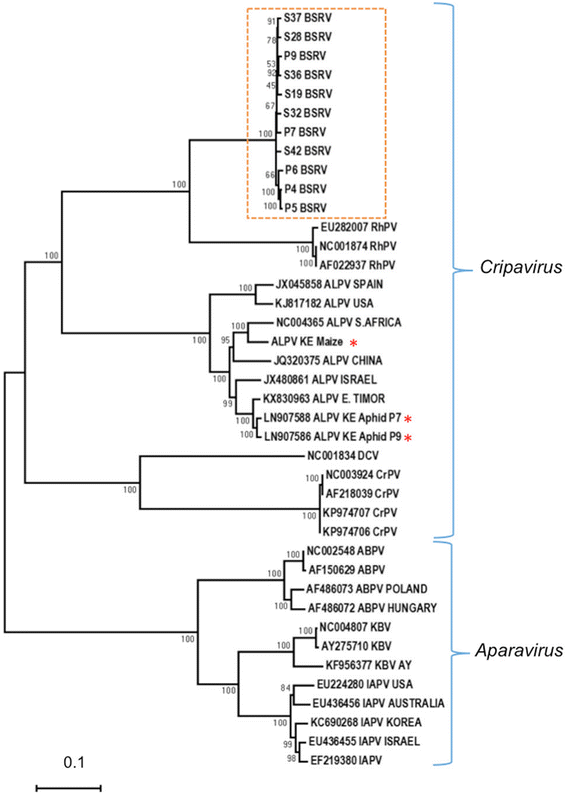
Results: Cytochrome oxidase 1 (CO1) gene sequencing showed that Aphis fabae was the sole aphid species present in bean plots in the farms visited. Sequencing of total RNA from aphids using the Illumina platform detected three dicistroviruses. Maize leaf RNA was also analysed. Identification of Aphid lethal paralysis virus (ALPV), Rhopalosiphum padi virus (RhPV), and a novel Big Sioux River virus (BSRV)-like dicistrovirus in aphid and maize samples was confirmed using reverse transcription-polymerase chain reactions and sequencing of amplified DNA products. Phylogenetic, nucleotide and protein sequence analyses of eight ALPV genomes revealed evidence of intra-species recombination, with the data suggesting there may be two ALPV lineages. Analysis of BSRV-like virus genomic RNA sequences revealed features that are consistent with other dicistroviruses and that it is phylogenetically closely related to dicistroviruses of the genus Cripavirus.
Conclusions: The discovery of ALPV and RhPV in aphids and maize further demonstrates the broad occurrence of these dicistroviruses. Dicistroviruses are remarkable in that they use plants as reservoirs that facilitate infection of their insect replicative hosts, such as aphids. This is the first report of these viruses being isolated from either organism. The BSRV-like sequences represent a potentially novel dicistrovirus infecting A. fabae.
Keywords: Aphid; Dicistrovirus; Epidemiology; Metagenomics; Phylogenetics; Potyvirus; Recombination; Vector.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures



Similar articles
-
Next-Generation Sequencing on Insectivorous Bat Guano: An Accurate Tool to Identify Arthropod Viruses of Potential Agricultural Concern.Viruses. 2019 Nov 28;11(12):1102. doi: 10.3390/v11121102. Viruses. 2019. PMID: 31795197 Free PMC article.
-
Analysis of new aphid lethal paralysis virus (ALPV) isolates suggests evolution of two ALPV species.J Gen Virol. 2014 Dec;95(Pt 12):2809-2819. doi: 10.1099/vir.0.069765-0. Epub 2014 Aug 28. J Gen Virol. 2014. PMID: 25170050
-
A metagenomic study of DNA viruses from samples of local varieties of common bean in Kenya.PeerJ. 2019 Mar 15;7:e6465. doi: 10.7717/peerj.6465. eCollection 2019. PeerJ. 2019. PMID: 30891366 Free PMC article.
-
Aphid Transmission of Potyvirus: The Largest Plant-Infecting RNA Virus Genus.Viruses. 2020 Jul 17;12(7):773. doi: 10.3390/v12070773. Viruses. 2020. PMID: 32708998 Free PMC article. Review.
-
Dicistroviruses.Annu Rev Entomol. 2010;55:129-50. doi: 10.1146/annurev-ento-112408-085457. Annu Rev Entomol. 2010. PMID: 19961327 Review.
Cited by
-
Virome Analysis of Aphid Populations That Infest the Barley Field: The Discovery of Two Novel Groups of Nege/Kita-Like Viruses and Other Novel RNA Viruses.Front Microbiol. 2020 Apr 7;11:509. doi: 10.3389/fmicb.2020.00509. eCollection 2020. Front Microbiol. 2020. PMID: 32318034 Free PMC article.
-
Post-COVID-19 Action: Guarding Africa's Crops against Viral Epidemics Requires Research Capacity Building That Unifies a Trio of Transdisciplinary Interventions.Viruses. 2020 Nov 9;12(11):1276. doi: 10.3390/v12111276. Viruses. 2020. PMID: 33182262 Free PMC article.
-
Untangling an insect's virome from its endogenous viral elements.BMC Genomics. 2023 Oct 24;24(1):636. doi: 10.1186/s12864-023-09737-z. BMC Genomics. 2023. PMID: 37875824 Free PMC article.
-
Metagenomic Analysis of Plant Virus Occurrence in Common Bean (Phaseolus vulgaris) in Central Kenya.Front Microbiol. 2018 Dec 7;9:2939. doi: 10.3389/fmicb.2018.02939. eCollection 2018. Front Microbiol. 2018. PMID: 30581419 Free PMC article.
-
Wisteria Vein Mosaic Virus Detected for the First Time in Iran from an Unknown Host by Analysis of Aphid Vectors.Plant Pathol J. 2020 Feb;36(1):87-97. doi: 10.5423/PPJ.OA.10.2019.0268. Epub 2020 Feb 1. Plant Pathol J. 2020. PMID: 32089664 Free PMC article.
References
-
- Christian PD, Scotti PD. Biopesticides from small RNA viruses on insects: aspects of their in vitro production. In: Maramorosch K, Loeb MJ, editors. Invertebrate cell culture: looking toward the twenty first century. San Francisco: Proc. Soc. in vitro Biol; 1996. pp. 73–81.
-
- Gildow FE, D’Arcy CJ. Barley and oats for an aphid virus and the influence on barley yellow dwarf virus transmission. Phytopathology. 1988;78:811–816. doi: 10.1094/Phyto-78-811. - DOI
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

