Massive A-to-I RNA editing is common across the Metazoa and correlates with dsRNA abundance
- PMID: 28969707
- PMCID: PMC5625713
- DOI: 10.1186/s13059-017-1315-y
Massive A-to-I RNA editing is common across the Metazoa and correlates with dsRNA abundance
Abstract
Background: Adenosine to inosine (A-to-I) RNA editing is a post-transcriptional modification catalyzed by the ADAR (adenosine deaminase that acts on RNA) enzymes, which are ubiquitously expressed among metazoans. Technical requirements have limited systematic mapping of editing sites to a small number of organisms. Thus, the extent of editing across the metazoan lineage is largely unknown.
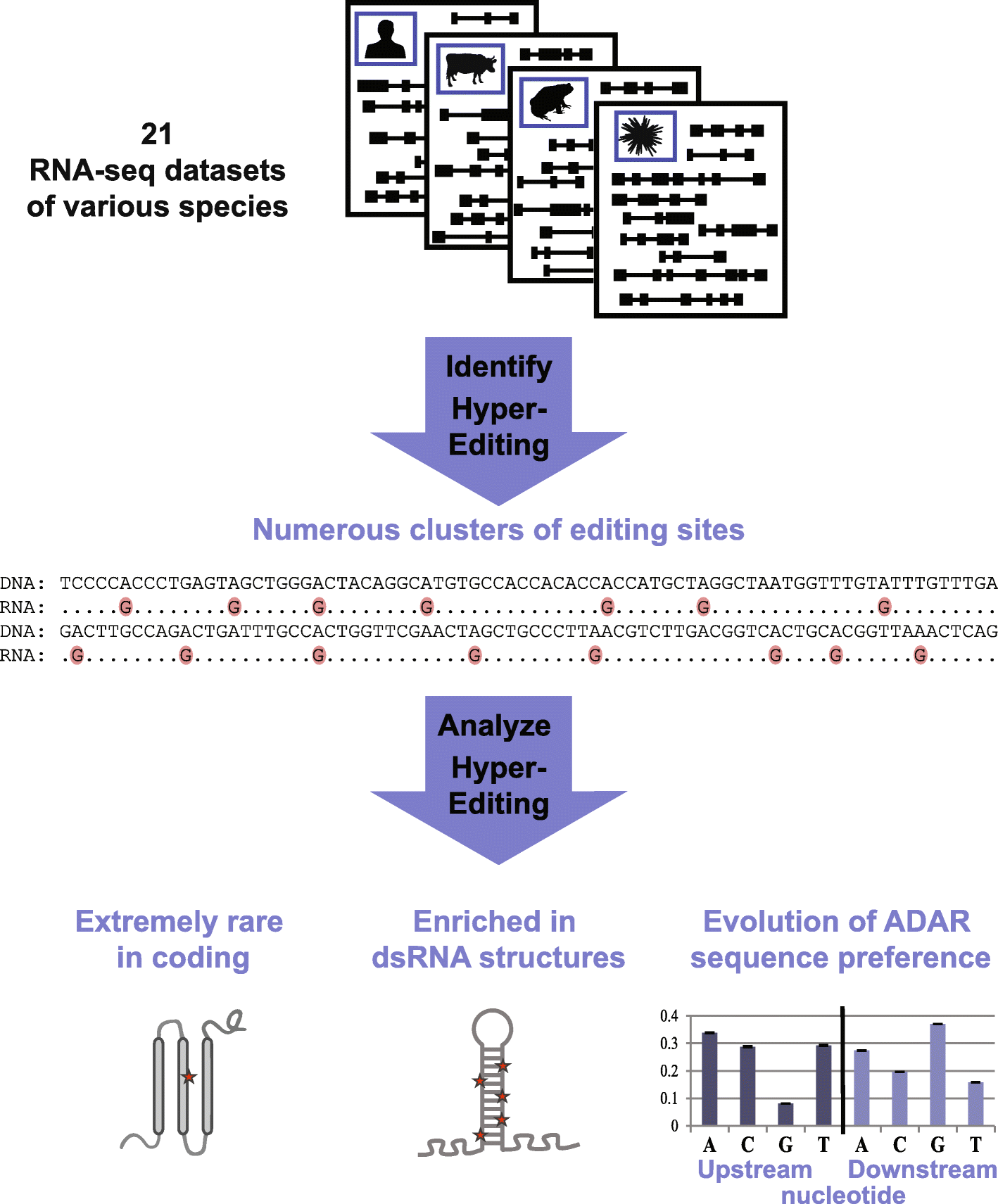
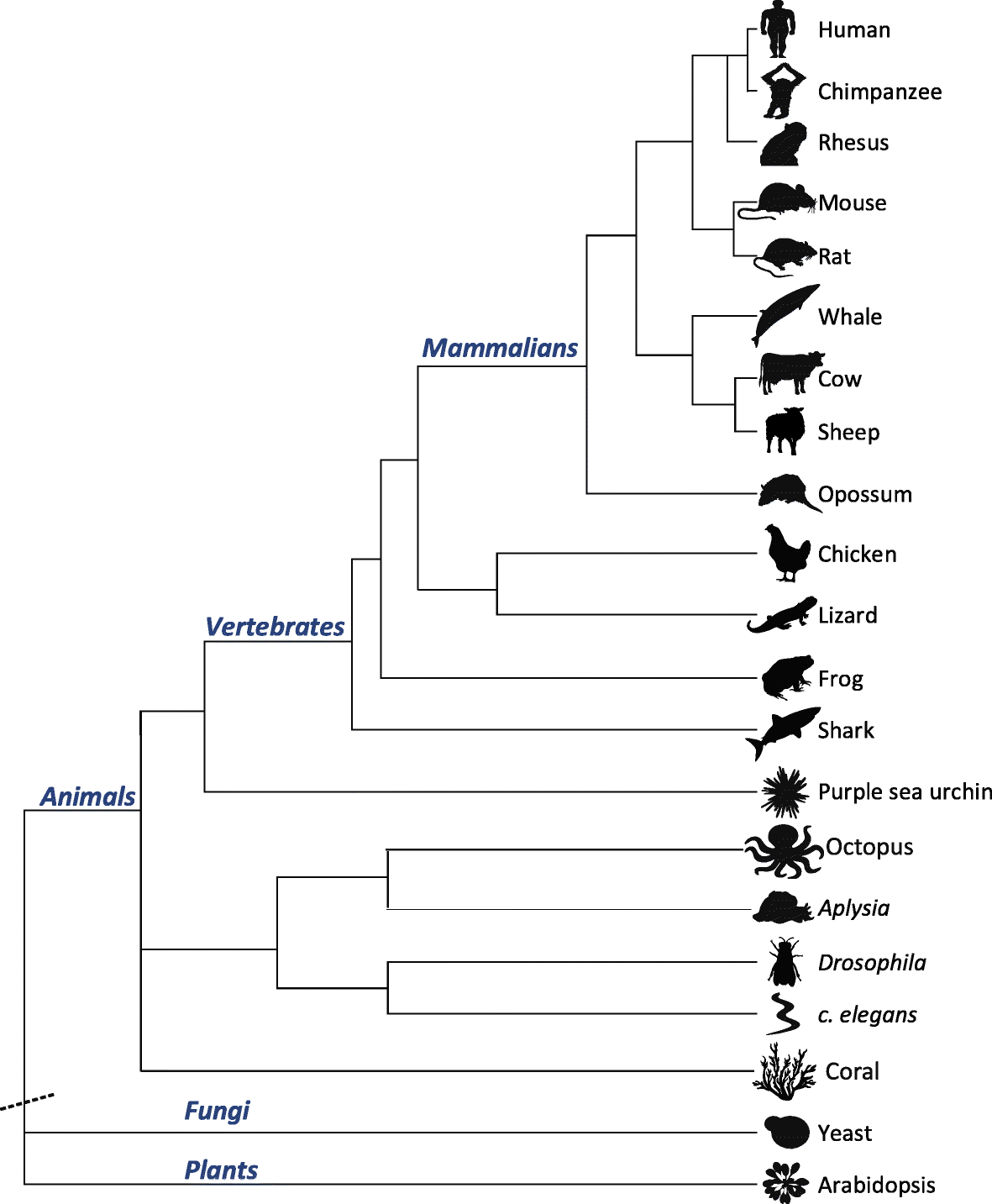
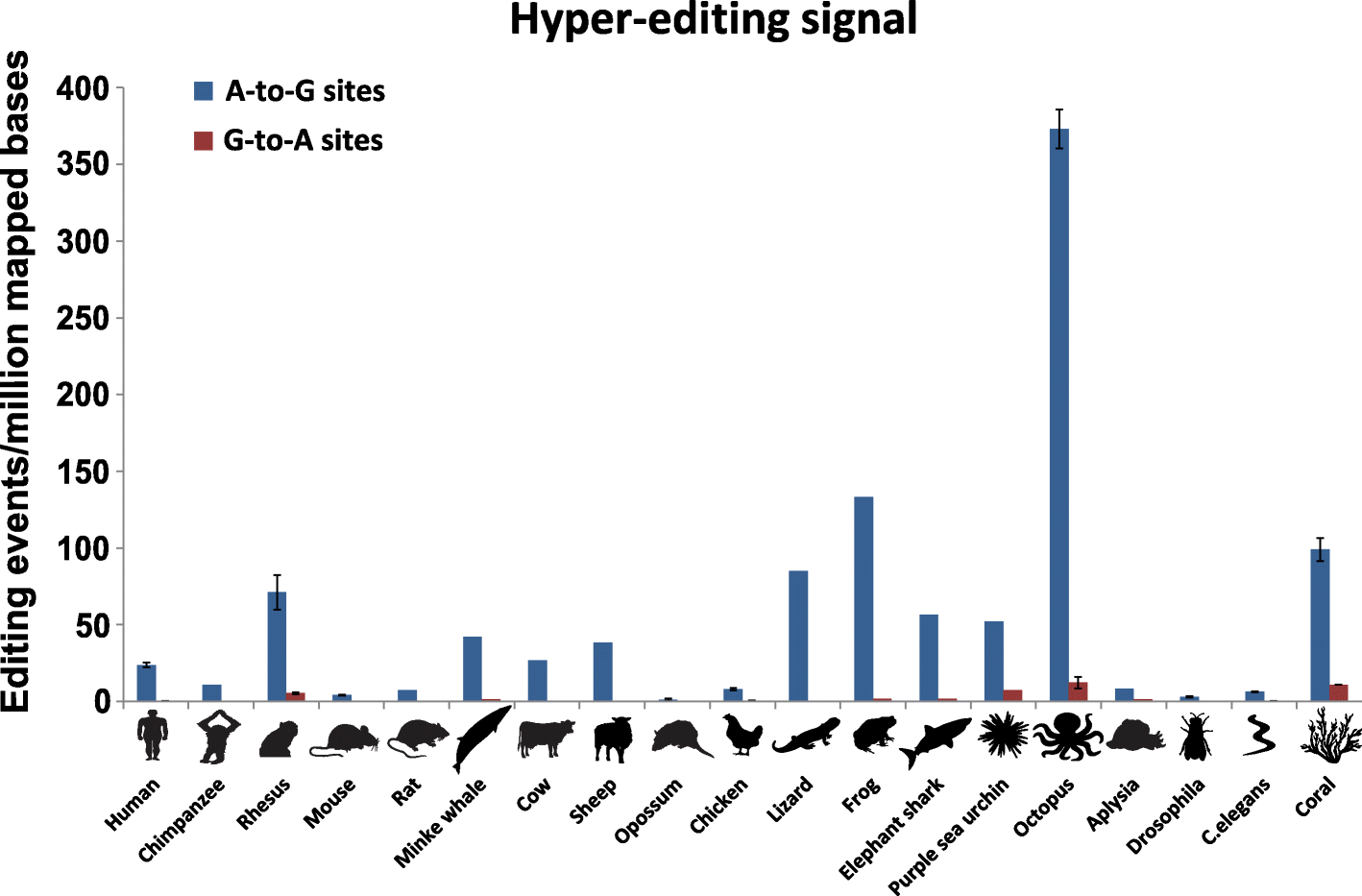
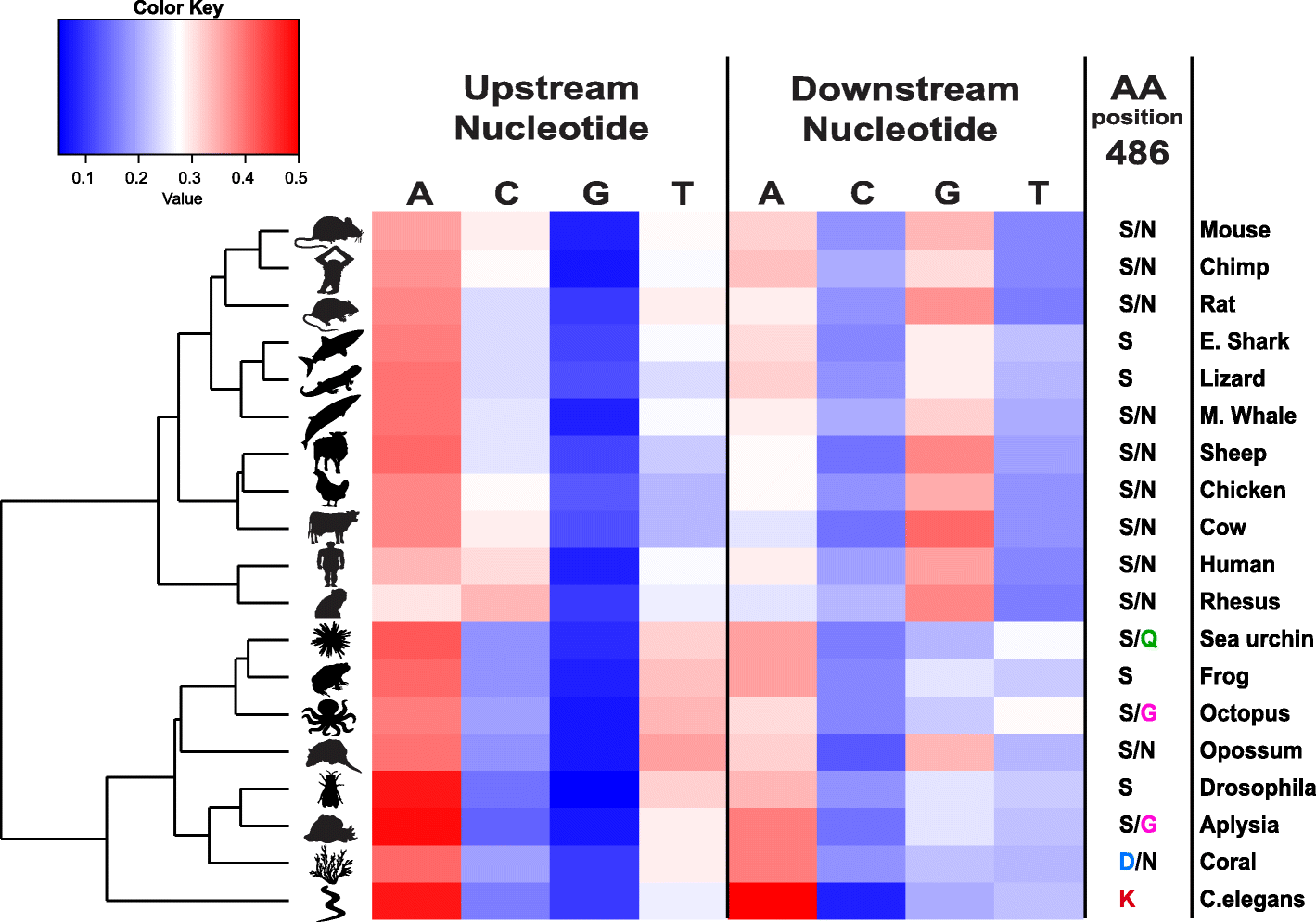
Results: Here, we apply a computational procedure to search for RNA-sequencing reads containing clusters of editing sites in 21 diverse organisms. Clusters of editing sites are abundant in repetitive genomic regions that putatively form double-stranded RNA (dsRNA) structures and are rarely seen in coding regions. The method reveals a considerable variation in hyper-editing levels across species, which is partly explained by differences in the potential of sequences to form dsRNA structures and the variability of ADAR proteins. Several commonly used model animals exhibit low editing levels and editing levels in primates is not exceptionally high, as previously suggested.
Conclusions: Editing by ADARs is highly prevalent across the Metazoa, mostly targeting dsRNA structures formed by genomic repeats. The degree to which the transcriptome of a given species undergoes hyper-editing is governed by the repertoire of repeats in the underlying genome. The strong association of RNA editing with the long dsRNA regions originating from non-coding repetitive elements is contrasted by the almost non-existing signal seen in coding regions. Hyper-edited regions are rarely expressed in a non-edited form. These results support the notion that the main role of ADAR is to suppress the cellular response to endogenous dsRNA structures.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

