Human Kidney Tubule-Specific Gene Expression Based Dissection of Chronic Kidney Disease Traits
- PMID: 28970079
- PMCID: PMC5652292
- DOI: 10.1016/j.ebiom.2017.09.014
Human Kidney Tubule-Specific Gene Expression Based Dissection of Chronic Kidney Disease Traits
Abstract
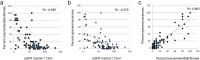
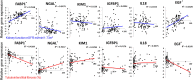
Chronic kidney disease (CKD) has diverse phenotypic manifestations including structural (such as fibrosis) and functional (such as glomerular filtration rate and albuminuria) alterations. Gene expression profiling has recently gained popularity as an important new tool for precision medicine approaches. Here we used unbiased and directed approaches to understand how gene expression captures different CKD manifestations in patients with diabetic and hypertensive CKD. Transcriptome data from ninety-five microdissected human kidney samples with a range of demographics, functional and structural changes were used for the primary analysis. Data obtained from 41 samples were available for validation. Using the unbiased Weighted Gene Co-Expression Network Analysis (WGCNA) we identified 16 co-expressed gene modules. We found that modules that strongly correlated with eGFR primarily encoded genes with metabolic functions. Gene groups that mainly encoded T-cell receptor and collagen pathways, showed the strongest correlation with fibrosis level, suggesting that these two phenotypic manifestations might have different underlying mechanisms. Linear regression models were then used to identify genes whose expression showed significant correlation with either structural (fibrosis) or functional (eGFR) manifestation and mostly corroborated the WGCNA findings. We concluded that gene expression is a very sensitive sensor of fibrosis, as the expression of 1654 genes correlated with fibrosis even after adjusting to eGFR and other clinical parameters. The association between GFR and gene expression was mostly mediated by fibrosis. In conclusion, our transcriptome-based CKD trait dissection analysis suggests that the association between gene expression and renal function is mediated by structural changes and that there may be differences in pathways that lead to decline in kidney function and the development of fibrosis, respectively.
Keywords: CKD; Fibrosis; Gene expression.
Copyright © 2017 The Authors. Published by Elsevier B.V. All rights reserved.
Figures





References
-
- An Y., Xu F., Le W., Ge Y., Zhou M., Chen H., Zeng C., Zhang H., Liu Z. Renal histologic changes and the outcome in patients with diabetic nephropathy. Nephrol. Dial. Transplant. 2015;30:257–266. - PubMed
-
- Becker G.J., Hewitson T.D. Animal models of chronic kidney disease: useful but not perfect. Nephrol. Dial. Transplant. 2013;28:2432–2438. - PubMed
-
- Bohle A., Mackensen-Haen S., Von Gise H., Grund K.E., Wehrmann M., Batz C., Bogenschutz O., Schmitt H., Nagy J., Muller C. The consequences of tubulo-interstitial changes for renal function in glomerulopathies. A morphometric and cytological analysis. Pathol. Res. Pract. 1990;186:135–144. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

