Synergistic Effect in Core Microbiota Associated with Sulfur Metabolism in Spontaneous Chinese Liquor Fermentation
- PMID: 28970229
- PMCID: PMC5717201
- DOI: 10.1128/AEM.01475-17
Synergistic Effect in Core Microbiota Associated with Sulfur Metabolism in Spontaneous Chinese Liquor Fermentation
Abstract
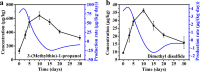
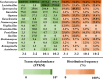
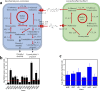
Microbial sulfur metabolism plays crucial roles in various food and alcoholic beverage fermentations. 3-(Methylthio)-1-propanol and dimethyl disulfide are important sulfur compounds in fermented foods and alcoholic beverages. Here, we studied the dynamics of these two compounds during spontaneous Chinese liquor fermentation. The two compounds reached the maximum concentration at day 10 and the maximum production rate at day 3. Metatranscriptomic analysis at days 3 and 10 revealed a total of 354 metabolically active microorganisms. Saccharomyces and Lactobacillus were identified as core microbiota critical for sulfur compound production based on both the transcript abundances of the principal genes and the distribution frequencies of 31 enzymes involved in sulfur metabolism. Saccharomyces transcribed genes encoding 23 enzymes related to the generation of 3-(methylthio)-1-propanol and dimethyl disulfide, and Lactobacillus was active in the methyl cycle, which recycles methionine, the precursor of the two sulfur compounds. Furthermore, the sulfur metabolism-related characteristics of two representative species were studied in coculture during a simulated fermentation. Saccharomyces cerevisiae JZ109 produced 158.4 μg/liter 3-(methylthio)-1-propanol and 58.5 μg/liter dimethyl disulfide in monoculture, whereas Lactobacillus buchneri JZ-JN-2017 could not produce these two compounds in monoculture. Their coculture significantly enhanced the generation of 3-(methylthio)-1-propanol (350.0 μg/liter) and dimethyl disulfide (123.8 μg/liter). In addition, coculture significantly enhanced the gene transcriptions (fold change, 1.5 to ∼55.0) that convert methionine to these two compounds in S. cerevisiae and in the methyl cycle of L. buchneri This study reveals a novel synergistic effect between members of the core microbiota in the production of sulfur compounds via methionine recycling in spontaneous Chinese liquor fermentation.IMPORTANCE Sulfur compounds play a crucial role in the aroma quality of various fermented foods and alcoholic beverages. However, it is unclear how these compounds are produced by microbes during their spontaneous fermentations. Here, we identified the core microbiota (Saccharomyces and Lactobacillus) associated with sulfur metabolism by determining both transcript abundance and distribution frequency of each genus in spontaneous Chinese liquor fermentation. This study provides a system-level analysis of sulfur metabolism by the metatranscriptomic analysis and culture-dependent methods. It sheds new light on how the metabolic behavior of the microbiota contributes to the liquor aroma quality. Furthermore, this work reveals a novel synergistic effect between Saccharomyces and Lactobacillus in the production of sulfur compounds, in which Lactobacillus regenerates the precursor methionine for sulfur compound production by Saccharomyces Our findings can contribute to the enhancement of aroma characteristics in Chinese liquor and open new avenues for improving various food and alcoholic beverage fermentation processes.
Keywords: Chinese liquor; core microbiota; metatranscriptomic; microbial interaction; sulfur metabolism; synergistic effect.
Copyright © 2017 American Society for Microbiology.
Figures



Similar articles
-
Construction of Synthetic Microbiota for Reproducible Flavor Compound Metabolism in Chinese Light-Aroma-Type Liquor Produced by Solid-State Fermentation.Appl Environ Microbiol. 2019 May 2;85(10):e03090-18. doi: 10.1128/AEM.03090-18. Print 2019 May 15. Appl Environ Microbiol. 2019. PMID: 30850432 Free PMC article.
-
Initial acidity regulates microbial sulfur metabolism in the spontaneous fermentation of sesame flavor-type baijiu.Int J Food Microbiol. 2025 May 2;435:111182. doi: 10.1016/j.ijfoodmicro.2025.111182. Epub 2025 Mar 31. Int J Food Microbiol. 2025. PMID: 40168753
-
Chinese Liquor Fermentation: Identification of Key Flavor-Producing Lactobacillus spp. by Quantitative Profiling with Indigenous Internal Standards.Appl Environ Microbiol. 2020 Jun 2;86(12):e00456-20. doi: 10.1128/AEM.00456-20. Print 2020 Jun 2. Appl Environ Microbiol. 2020. PMID: 32276974 Free PMC article.
-
Research Progress on Flavor Compounds and Microorganisms of Maotai Flavor Baijiu.J Food Sci. 2019 Jan;84(1):6-18. doi: 10.1111/1750-3841.14409. Epub 2018 Dec 12. J Food Sci. 2019. PMID: 30548499 Review.
-
Occurrence and taxonomic characteristics of strains of Saccharomyces cerevisiae predominant in African indigenous fermented foods and beverages.FEMS Yeast Res. 2003 Apr;3(2):191-200. doi: 10.1016/S1567-1356(02)00185-X. FEMS Yeast Res. 2003. PMID: 12702452 Review.
Cited by
-
Compositional Differences and Similarities between Typical Chinese Baijiu and Western Liquor as Revealed by Mass Spectrometry-Based Metabolomics.Metabolites. 2018 Dec 21;9(1):2. doi: 10.3390/metabo9010002. Metabolites. 2018. PMID: 30577624 Free PMC article.
-
Metabolic cooperation between conspecific genotypic groups contributes to bacterial fitness.ISME Commun. 2023 Apr 28;3(1):41. doi: 10.1038/s43705-023-00250-8. ISME Commun. 2023. PMID: 37117489 Free PMC article.
-
Isolation and Characterization of Yeast with Benzenemethanethiol Synthesis Ability Isolated from Baijiu Daqu.Foods. 2023 Jun 23;12(13):2464. doi: 10.3390/foods12132464. Foods. 2023. PMID: 37444202 Free PMC article.
-
Effect of Yeast Inoculation on the Bacterial Community Structure in Reduced-Salt Harbin Dry Sausages: A Perspective of Fungi-Bacteria Interactions.Foods. 2024 Jan 18;13(2):307. doi: 10.3390/foods13020307. Foods. 2024. PMID: 38254608 Free PMC article.
-
Specific Volumetric Weight-Driven Shift in Microbiota Compositions With Saccharifying Activity Change in Starter for Chinese Baijiu Fermentation.Front Microbiol. 2018 Sep 28;9:2349. doi: 10.3389/fmicb.2018.02349. eCollection 2018. Front Microbiol. 2018. PMID: 30323805 Free PMC article.
References
-
- Jin G, Zhu Y, Xu Y. 2017. Mystery behind Chinese liquor fermentation. Trends Food Sci Technol 63:18–28. doi:10.1016/j.tifs.2017.02.016. - DOI
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

