Using chirality to probe the conformational dynamics and assembly of intrinsically disordered amyloid proteins
- PMID: 28970487
- PMCID: PMC5624888
- DOI: 10.1038/s41598-017-10525-5
Using chirality to probe the conformational dynamics and assembly of intrinsically disordered amyloid proteins
Abstract
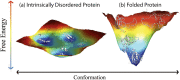
Intrinsically disordered protein (IDP) conformers occupy large regions of conformational space and display relatively flat energy surfaces. Amyloid-forming IDPs, unlike natively folded proteins, have folding trajectories that frequently involve movements up shallow energy gradients prior to the "downhill" folding leading to fibril formation. We suggest that structural perturbations caused by chiral inversions of amino acid side-chains may be especially valuable in elucidating these pathways of IDP folding. Chiral inversions are subtle in that they do not change side-chain size, flexibility, hydropathy, charge, or polarizability. They allow focus to be placed solely on the question of how changes in amino acid side-chain orientation, and the resultant alterations in peptide backbone structure, affect a peptide's conformational landscape (Ramachandran space). If specific inversions affect folding and assembly, then the sites involved likely are important in mediating these processes. We suggest here a "focused chiral mutant library" approach for the unbiased study of amyloid-forming IDPs.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

Similar articles
-
Energy Landscapes of Protein Aggregation and Conformation Switching in Intrinsically Disordered Proteins.J Mol Biol. 2021 Oct 1;433(20):167182. doi: 10.1016/j.jmb.2021.167182. Epub 2021 Aug 3. J Mol Biol. 2021. PMID: 34358545 Review.
-
The Thermodynamic Basis of the Fuzzy Interaction of an Intrinsically Disordered Protein.Angew Chem Int Ed Engl. 2017 Nov 13;56(46):14494-14497. doi: 10.1002/anie.201707853. Epub 2017 Oct 10. Angew Chem Int Ed Engl. 2017. PMID: 28914483
-
Complete Coupled Binding-Folding Pathway of the Intrinsically Disordered Transcription Factor Protein Brinker Revealed by Molecular Dynamics Simulations and Markov State Modeling.Biochemistry. 2018 Jul 31;57(30):4404-4420. doi: 10.1021/acs.biochem.8b00441. Epub 2018 Jul 19. Biochemistry. 2018. PMID: 29990433
-
Formation of Heterotypic Amyloids: α-Synuclein in Co-Aggregation.Proteomics. 2018 Nov;18(21-22):e1800059. doi: 10.1002/pmic.201800059. Epub 2018 Oct 10. Proteomics. 2018. PMID: 30216674 Review.
-
Cross-Linking Mass Spectrometry Analysis of Metastable Compact Structures in Intrinsically Disordered Proteins.Methods Mol Biol. 2023;2551:189-201. doi: 10.1007/978-1-0716-2597-2_13. Methods Mol Biol. 2023. PMID: 36310204
Cited by
-
Monitoring α-synuclein aggregation.Neurobiol Dis. 2023 Jan;176:105966. doi: 10.1016/j.nbd.2022.105966. Epub 2022 Dec 15. Neurobiol Dis. 2023. PMID: 36527982 Free PMC article. Review.
-
Stereoselectivity of Interaction of Nonsteroidal Anti-Inflammatory Drug S-Ketoprofen with L/D-Tryptophan in Phospholipid Membranes.Membranes (Basel). 2022 Apr 24;12(5):460. doi: 10.3390/membranes12050460. Membranes (Basel). 2022. PMID: 35629787 Free PMC article.
-
Insights and Perspectives on the Role of Proteostasis and Heat Shock Proteins in Fungal Infections.Microorganisms. 2023 Jul 25;11(8):1878. doi: 10.3390/microorganisms11081878. Microorganisms. 2023. PMID: 37630438 Free PMC article. Review.
-
Role of Chiral Configuration in the Photoinduced Interaction of D- and L-Tryptophan with Optical Isomers of Ketoprofen in Linked Systems.Int J Mol Sci. 2021 Jun 8;22(12):6198. doi: 10.3390/ijms22126198. Int J Mol Sci. 2021. PMID: 34201293 Free PMC article.
-
Understanding and controlling amyloid aggregation with chirality.Curr Opin Chem Biol. 2021 Oct;64:1-9. doi: 10.1016/j.cbpa.2021.01.003. Epub 2021 Feb 18. Curr Opin Chem Biol. 2021. PMID: 33610939 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

