The FXR Agonist, Obeticholic Acid, Suppresses HCC Proliferation & Metastasis: Role of IL-6/STAT3 Signalling Pathway
- PMID: 28970500
- PMCID: PMC5624958
- DOI: 10.1038/s41598-017-12629-4
The FXR Agonist, Obeticholic Acid, Suppresses HCC Proliferation & Metastasis: Role of IL-6/STAT3 Signalling Pathway
Abstract
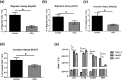
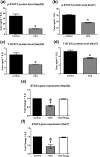
The nuclear receptor, farnesoid X receptor (FXR), has been recently considered as a tumor suppressor in HCC. IL-6/Janus kinase 2 (Jak-2)/signal transducer and activator of transcription 3 (STAT3) pathway has been implicated as a key player in many cancer types. This study aimed at investigating the potential effect of the FXR agonist, obeticholic acid (OCA), on HCC and the involvement of IL-6/STAT3 pathway. The potential regulation of STAT3 by its main feedback inhibitor target gene, the suppressor of cytokine signaling 3 (SOCS3), triggered by OCA was also explored. Cytotoxicity studies were performed on HepG2, Huh7, and SNU-449 cell lines using OCA alone and combined with the FXR antagonist guggulsterone (Gugg). OCA cytotoxic effect was significantly hampered in presence of Gugg. OCA also caused cell cycle arrest and inhibited invasion and migration of HCC cells. Decrease in STAT3 phosphorylation and SOCS3 upregulation were also observed. Moreover, Jak-2, IL-1β, and IL-6 levels were decreased. These results were correlated with an upregulation of FXR and small heterodimer partner (SHP) levels. Effects of OCA on IL-6/STAT3 main key players were reversed in presence of Gugg. Overall, these findings suggest a potential effect of OCA in HCC via interfering with IL-6/STAT3 signalling pathway in vitro.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

