Transcriptomes and neurotransmitter profiles of classes of gustatory and somatosensory neurons in the geniculate ganglion
- PMID: 28970527
- PMCID: PMC5624912
- DOI: 10.1038/s41467-017-01095-1
Transcriptomes and neurotransmitter profiles of classes of gustatory and somatosensory neurons in the geniculate ganglion
Abstract
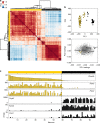
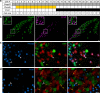
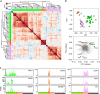
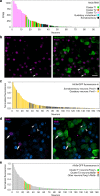
Taste buds are innervated by neurons whose cell bodies reside in cranial sensory ganglia. Studies on the functional properties and connectivity of these neurons are hindered by the lack of markers to define their molecular identities and classes. The mouse geniculate ganglion contains chemosensory neurons innervating lingual and palatal taste buds and somatosensory neurons innervating the pinna. Here, we report single cell RNA sequencing of geniculate ganglion neurons. Using unbiased transcriptome analyses, we show a pronounced separation between two major clusters which, by anterograde labeling, correspond to gustatory and somatosensory neurons. Among the gustatory neurons, three subclusters are present, each with its own complement of transcription factors and neurotransmitter response profiles. The smallest subcluster expresses both gustatory- and mechanosensory-related genes, suggesting a novel type of sensory neuron. We identify several markers to help dissect the functional distinctions among gustatory neurons and address questions regarding target interactions and taste coding.Characterization of gustatory neural pathways has suffered due to a lack of molecular markers. Here, the authors report single cell RNA sequencing and unbiased transcriptome analyses to reveal major distinctions between gustatory and somatosensory neurons and subclusters of gustatory neurons with unique molecular and functional profiles.
Conflict of interest statement
The authors declare that there are no competing financial interests.
Figures



Similar articles
-
The transcription factor Phox2b distinguishes between oral and non-oral sensory neurons in the geniculate ganglion.J Comp Neurol. 2017 Dec 15;525(18):3935-3950. doi: 10.1002/cne.24312. Epub 2017 Sep 15. J Comp Neurol. 2017. PMID: 28856690 Free PMC article.
-
EGR4 is critical for cell-fate determination and phenotypic maintenance of geniculate ganglion neurons underlying sweet and umami taste.Proc Natl Acad Sci U S A. 2023 May 30;120(22):e2217595120. doi: 10.1073/pnas.2217595120. Epub 2023 May 22. Proc Natl Acad Sci U S A. 2023. PMID: 37216536 Free PMC article.
-
Oral Sensory Neurons of the Geniculate Ganglion That Express Tyrosine Hydroxylase Comprise a Subpopulation That Contacts Type II and Type III Taste Bud Cells.eNeuro. 2022 Oct 13;9(5):ENEURO.0523-21.2022. doi: 10.1523/ENEURO.0523-21.2022. Print 2022 Sep-Oct. eNeuro. 2022. PMID: 36216506 Free PMC article.
-
Building sensory receptors on the tongue.J Neurocytol. 2004 Dec;33(6):631-46. doi: 10.1007/s11068-005-3332-0. Epub 2005 Oct 11. J Neurocytol. 2004. PMID: 16217619 Review.
-
Advances in Optical Tools to Study Taste Sensation.Mol Cells. 2022 Dec 31;45(12):877-882. doi: 10.14348/molcells.2022.0116. Epub 2022 Dec 13. Mol Cells. 2022. PMID: 36572557 Free PMC article. Review.
Cited by
-
Selectively Imaging Cranial Sensory Ganglion Neurons Using AAV-PHP.S.eNeuro. 2022 Jun 6;9(3):ENEURO.0373-21.2022. doi: 10.1523/ENEURO.0373-21.2022. Print 2022 May-Jun. eNeuro. 2022. PMID: 35610024 Free PMC article.
-
COVID-19 and the Chemical Senses: Supporting Players Take Center Stage.Neuron. 2020 Jul 22;107(2):219-233. doi: 10.1016/j.neuron.2020.06.032. Epub 2020 Jul 1. Neuron. 2020. PMID: 32640192 Free PMC article. Review.
-
Microfluidics for Neuronal Cell and Circuit Engineering.Chem Rev. 2022 Sep 28;122(18):14842-14880. doi: 10.1021/acs.chemrev.2c00212. Epub 2022 Sep 7. Chem Rev. 2022. PMID: 36070858 Free PMC article. Review.
-
Anterior and Posterior Tongue Regions and Taste Papillae: Distinct Roles and Regulatory Mechanisms with an Emphasis on Hedgehog Signaling and Antagonism.Int J Mol Sci. 2023 Mar 2;24(5):4833. doi: 10.3390/ijms24054833. Int J Mol Sci. 2023. PMID: 36902260 Free PMC article. Review.
-
Function, Innervation, and Neurotransmitter Signaling in Mice Lacking Type-II Taste Cells.eNeuro. 2020 Feb 3;7(1):ENEURO.0339-19.2020. doi: 10.1523/ENEURO.0339-19.2020. Print 2020 Jan/Feb. eNeuro. 2020. PMID: 31988217 Free PMC article.
References
-
- Lundy RF, Jr., Contreras RJ. Gustatory neuron types in rat geniculate ganglion. J. Neurophysiol. 1999;82:2970–2988. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

