Comprehensive Analysis of DWARF14-LIKE2 (DLK2) Reveals Its Functional Divergence from Strigolactone-Related Paralogs
- PMID: 28970845
- PMCID: PMC5609103
- DOI: 10.3389/fpls.2017.01641
Comprehensive Analysis of DWARF14-LIKE2 (DLK2) Reveals Its Functional Divergence from Strigolactone-Related Paralogs
Abstract
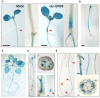
Strigolactones (SLs) and related butenolides, originally identified as active seed germination stimulants of parasitic weeds, play important roles in many aspects of plant development. Two members of the D14 α/β hydrolase protein family, DWARF14 (D14) and KARRIKIN INSENSITIVE2 (KAI2) are essential for SL/butenolide signaling. The third member of the family in Arabidopsis, DWARF 14-LIKE2 (DLK2) is structurally very similar to D14 and KAI2, but its function is unknown. We demonstrated that DLK2 does not bind nor hydrolyze natural (+)5-deoxystrigol [(+)5DS], and weakly hydrolyzes non-natural strigolactone (-)5DS. A detailed genetic analysis revealed that DLK2 does not affect SL responses and can regulate seedling photomorphogenesis. DLK2 is upregulated in the dark dependent upon KAI2 and PHYTOCHROME INTERACTING FACTORS (PIFs), indicating that DLK2 might function in light signaling pathways. In addition, unlike its paralog proteins, DLK2 is not subject to rac-GR24-induced degradation, suggesting that DLK2 acts independently of MORE AXILLARY GROWTH2 (MAX2); however, regulation of DLK2 transcription is mostly accomplished through MAX2. In conclusion, these data suggest that DLK2 represents a divergent member of the DWARF14 family.
Keywords: AtD14; DLK2; KAI2; MAX2; butenolide; light; strigolactone.
Figures






References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

