Comparison of the Luminal and Mucosa-Associated Microbiota in the Colon of Pigs with and without Swine Dysentery
- PMID: 28971100
- PMCID: PMC5609587
- DOI: 10.3389/fvets.2017.00139
Comparison of the Luminal and Mucosa-Associated Microbiota in the Colon of Pigs with and without Swine Dysentery
Abstract
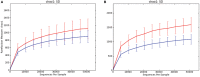
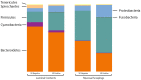
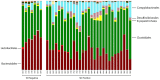
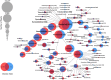
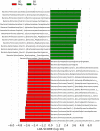
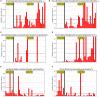
Colonic contents and mucosal scrapings from pigs inoculated with Brachyspira hyodysenteriae or Brachyspira hampsonii were collected at necropsy and classified as either positive (n = 29) or negative (n = 7) for swine dysentery (SD) based upon lesions and positive culture from the source pig. The microbiota in each sample was analyzed by bacterial census taking (16S rRNA gene sequencing). Procrustes analysis revealed similar clustering by disease classification with a relatively high M2 value (0.44) suggesting differences in the microbiota between mucosal and luminal samples from the same pig. In both sample types, differences in richness and beta diversity were observed between disease statuses (P ≤ 0.014). The relative abundance of Brachyspirales, Campylobacterales, Desulfovibrionales, and Enterobacteriales was higher in pigs with dysentery for both mucosal scrapings and luminal samples while Clostridiales, Erysipelotrichales, and Fusobacteriales were significantly more abundant in the luminal contents only. For inoculated pigs that did not develop dysentery, Burkholderiales were more abundant in both sample types, Bacteroidales and Synergistales were more abundant in mucosal scrapings, and Lactobacillales and Bifidobacteriales were more abundant in luminal contents when compared with diseased pigs. Linear discriminant analysis of effect size revealed Brachyspira, Campylobacter, Mogibacterium, and multiple Desulfovibrio spp. as differential features in mucosal scrapings from pigs with dysentery while Lactobacillus and a Bifidobacterium spp. were differential in pigs without disease. These differential features were not observed in luminal samples. In summary, microbial profiles in both sample types differ significantly between disease states; however, evaluation of the mucosal microbiome specifically may be of higher value in elucidating bacterial mechanisms underlying development of SD.
Keywords: Brachyspira hampsonii; Brachyspira hyodysenteriae; metagenomics; microbial profiling; swine; swine dysentery.
Figures
Similar articles
-
Comparison of sesion severity, distribution, and colonic mucin expression in pigs with acute swine dysentery following oral inoculation with "Brachyspira hampsonii" or Brachyspira hyodysenteriae.Vet Pathol. 2014 Nov;51(6):1096-108. doi: 10.1177/0300985813516646. Epub 2014 Feb 27. Vet Pathol. 2014. PMID: 24577722
-
Fluorescent in situ hybridization for detection of "Brachyspira hampsonii" in porcine colonic tissues.J Vet Diagn Invest. 2013 May;25(3):407-12. doi: 10.1177/1040638713485228. Epub 2013 Apr 9. J Vet Diagn Invest. 2013. PMID: 23572452
-
Comparative virulence of clinical Brachyspira spp. isolates in inoculated pigs.J Vet Diagn Invest. 2012 Nov;24(6):1025-34. doi: 10.1177/1040638712457927. Epub 2012 Sep 5. J Vet Diagn Invest. 2012. PMID: 22956484
-
Swine Dysentery.Vet Pathol. 2017 Jan;54(1):22-31. doi: 10.1177/0300985816653795. Epub 2016 Jul 11. Vet Pathol. 2017. PMID: 27288432 Review.
-
A review of methods used for studying the molecular epidemiology of Brachyspira hyodysenteriae.Vet Microbiol. 2017 Aug;207:181-194. doi: 10.1016/j.vetmic.2017.06.011. Epub 2017 Jun 19. Vet Microbiol. 2017. PMID: 28757022 Review.
Cited by
-
How Can We Define "Optimal Microbiota?": A Comparative Review of Structure and Functions of Microbiota of Animals, Fish, and Plants in Agriculture.Front Nutr. 2018 Oct 2;5:90. doi: 10.3389/fnut.2018.00090. eCollection 2018. Front Nutr. 2018. PMID: 30333981 Free PMC article. Review.
-
PCR-Based Analysis of ColE1 Plasmids in Clinical Isolates and Metagenomic Samples Reveals Their Importance as Gene Capture Platforms.Front Microbiol. 2018 Mar 16;9:469. doi: 10.3389/fmicb.2018.00469. eCollection 2018. Front Microbiol. 2018. PMID: 29615998 Free PMC article.
-
Commercial farmed swine harbour a variety of pathogenic bacteria and antimicrobial resistance genes.J Med Microbiol. 2024 Jan;73(1):001787. doi: 10.1099/jmm.0.001787. J Med Microbiol. 2024. PMID: 38230911 Free PMC article.
-
Assessing the Probiotic Effects of Pediococcus pentosaceus CACC616 in Weaned Piglets.Microorganisms. 2023 Nov 30;11(12):2890. doi: 10.3390/microorganisms11122890. Microorganisms. 2023. PMID: 38138034 Free PMC article.
-
Assessing the Impact of Diet on the Mucosa-Adhered Microbiome in Piglets Using Comparative Analysis of Rectal Swabs and Colon Content.Front Microbiol. 2022 Feb 22;13:804986. doi: 10.3389/fmicb.2022.804986. eCollection 2022. Front Microbiol. 2022. PMID: 35273582 Free PMC article.
References
-
- Hansen CF, Hernández A, Mansfield J, Hidalgo Á, La T, Phillips ND, et al. A high dietary concentration of inulin is necessary to reduce the incidence of swine dysentery in pigs experimentally challenged with Brachyspira hyodysenteriae. Br J Nutr (2011) 106:1506–13. 10.1017/S000711451100208X - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

