Philadelphia chromosome-like acute lymphoblastic leukemia
- PMID: 28972016
- PMCID: PMC5680607
- DOI: 10.1182/blood-2017-06-743252
Philadelphia chromosome-like acute lymphoblastic leukemia
Abstract
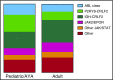
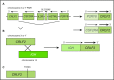
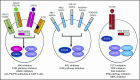
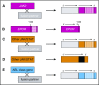
Philadelphia chromosome (Ph)-like acute lymphoblastic leukemia (ALL), also referred to as BCR-ABL1-like ALL, is a high-risk subset with a gene expression profile that shares significant overlap with that of Ph-positive (Ph+) ALL and is suggestive of activated kinase signaling. Although Ph+ ALL is defined by BCR-ABL1 fusion, Ph-like ALL cases contain a variety of genomic alterations that activate kinase and cytokine receptor signaling. These alterations can be grouped into major subclasses that include ABL-class fusions involving ABL1, ABL2, CSF1R, and PDGFRB that phenocopy BCR-ABL1 and alterations of CRLF2, JAK2, and EPOR that activate JAK/STAT signaling. Additional genomic alterations in Ph-like ALL activate other kinases, including BLNK, DGKH, FGFR1, IL2RB, LYN, NTRK3, PDGFRA, PTK2B, TYK2, and the RAS signaling pathway. Recent studies have helped to define the genomic landscape of Ph-like ALL and how it varies across the age spectrum, associated clinical features and outcomes, and genetic risk factors. Preclinical studies and anecdotal reports show that targeted inhibitors of relevant signaling pathways are active in specific Ph-like ALL subsets, and precision medicine trials have been initiated for this high-risk ALL subset.
© 2017 by The American Society of Hematology.
Figures

References
-
- Arber DA, Orazi A, Hasserjian R, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127(20):2391-2405. - PubMed
-
- Fielding AK. Treatment of Philadelphia chromosome-positive acute lymphoblastic leukemia in adults: a broader range of options, improved outcomes, and more therapeutic dilemmas. Am Soc Clin Oncol Educ Book. 2015;2015:e352-e359. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

