Zic-Proteins Are Repressors of Dopaminergic Forebrain Fate in Mice and C. elegans
- PMID: 28972122
- PMCID: PMC6596518
- DOI: 10.1523/JNEUROSCI.3888-16.2017
Zic-Proteins Are Repressors of Dopaminergic Forebrain Fate in Mice and C. elegans
Abstract
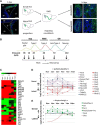
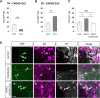
In the postnatal forebrain regionalized neural stem cells along the ventricular walls produce olfactory bulb (OB) interneurons with varying neurotransmitter phenotypes and positions. To understand the molecular basis of this region-specific variability we analyzed gene expression in the postnatal dorsal and lateral lineages in mice of both sexes from stem cells to neurons. We show that both lineages maintain transcription factor signatures of their embryonic site of origin, the pallium and subpallium. However, additional factors, including Zic1 and Zic2, are postnatally expressed in the dorsal stem cell compartment and maintained in the lineage that generates calretinin-positive GABAergic neurons for the OB. Functionally, we show that Zic1 and Zic2 induce the generation of calretinin-positive neurons while suppressing dopaminergic fate in the postnatal dorsal lineage. We investigated the evolutionary conservation of the dopaminergic repressor function of Zic proteins and show that it is already present in C. elegansSIGNIFICANCE STATEMENT The vertebrate brain generates thousands of different neuron types. In this work we investigate the molecular mechanisms underlying this variability. Using a genomics approach we identify the transcription factor signatures of defined neural stem cells and neuron populations. Based thereon we show that two related transcription factors, Zic1 and Zic2, are essential to control the balance between two defined neuron types in the postnatal brain. We show that this mechanism is conserved in evolutionary very distant species.
Keywords: neural stem cells; olfactory bulb; transcription factor.
Copyright © 2017 the authors 0270-6474/17/3710611-13$15.00/0.
Figures




References
-
- Alper S, Kenyon C (2002) The zinc finger protein REF-2 functions with the Hox genes to inhibit cell fusion in the ventral epidermis of C. elegans. Development 129:3335–3348. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
