Mutant cycle analysis identifies a ligand interaction site in an odorant receptor of the malaria vector Anopheles gambiae
- PMID: 28972152
- PMCID: PMC5704475
- DOI: 10.1074/jbc.M117.810374
Mutant cycle analysis identifies a ligand interaction site in an odorant receptor of the malaria vector Anopheles gambiae
Abstract
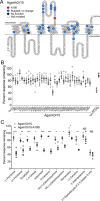
Lack of information about the structure of insect odorant receptors (ORs) hinders the development of more effective repellants to control disease-transmitting insects. Mutagenesis and functional analyses using agonists to map the odorant-binding sites of these receptors have been limited because mutations distant from an agonist-binding site can alter agonist sensitivity. Here we use mutant cycle analysis, an approach for exploring the energetics of protein-protein or protein-ligand interactions, with inhibitors, to identify a component of the odorant-binding site of an OR from the malaria vector, Anopheles gambiae The closely related odorant-specificity subunits Agam/Or15 and Agam/Or13 were each co-expressed with Agam/Orco (odorant receptor co-receptor subunit) in Xenopus oocytes and assayed by two-electrode voltage clamp electrophysiology. We identified (-)-fenchone as a competitive inhibitor with different potencies at the two receptors and used this difference to screen a panel of 37 Agam/Or15 mutants, surveying all positions that differ between Agam/Or15 and Agam/Or13 in the transmembrane and extracellular regions, identifying position 195 as a determinant of (-)-fenchone sensitivity. Inhibition by (-)-fenchone and six structurally related inhibitors of Agam/Or15 receptors containing each of four different hydrophobic residues at position 195 served as input data for mutant cycle analysis. Several mutant cycles, calculated from the inhibition of two receptors by each of two ligands, yielded coupling energies of ≥1 kcal/mol, indicating a close, physical interaction between the ligand and residue 195 of Agam/Or15. This approach should be useful in further expanding our knowledge of odorant-binding site structures in ORs of disease vector insects.
Keywords: Xenopus; electrophysiology; insect; olfaction; oocyte; receptor; structure.
© 2017 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures



References
-
- Guidobaldi F., May-Concha I. J., and Guerenstein P. G. (2014) Morphology and physiology of the olfactory system of blood-feeding insects. J. Physiol. Paris 108, 96–111 - PubMed
-
- Gibson G., and Torr S. J. (1999) Visual and olfactory responses of haematophagous Diptera to host stimuli. Med. Vet. Entomol. 13, 2–23 - PubMed
-
- Erdelyan C. N., Mahood T. H., Bader T. S., and Whyard S. (2012) Functional validation of the carbon dioxide receptor genes in Aedes aegypti mosquitoes using RNA interference. Insect Mol. Biol. 21, 119–127 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

