A conserved degron containing an amphipathic helix regulates the cholesterol-mediated turnover of human squalene monooxygenase, a rate-limiting enzyme in cholesterol synthesis
- PMID: 28972164
- PMCID: PMC5723984
- DOI: 10.1074/jbc.M117.794230
A conserved degron containing an amphipathic helix regulates the cholesterol-mediated turnover of human squalene monooxygenase, a rate-limiting enzyme in cholesterol synthesis
Abstract
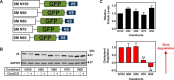
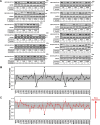
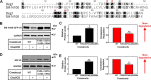
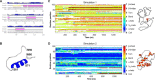
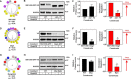
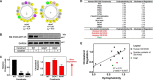
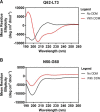
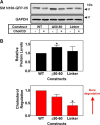
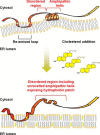
Cholesterol biosynthesis in the endoplasmic reticulum (ER) is tightly controlled by multiple mechanisms to regulate cellular cholesterol levels. Squalene monooxygenase (SM) is the second rate-limiting enzyme in cholesterol biosynthesis and is regulated both transcriptionally and post-translationally. SM undergoes cholesterol-dependent proteasomal degradation when cholesterol is in excess. The first 100 amino acids of SM (designated SM N100) are necessary for this degradative process and represent the shortest cholesterol-regulated degron identified to date. However, the fundamental intrinsic characteristics of this degron remain unknown. In this study, we performed a series of deletions, point mutations, and domain swaps to identify a 12-residue region (residues Gln-62-Leu-73), required for SM cholesterol-mediated turnover. Molecular dynamics and circular dichroism revealed an amphipathic helix within this 12-residue region. Moreover, 70% of the variation in cholesterol regulation was dependent on the hydrophobicity of this region. Of note, the earliest known Doa10 yeast degron, Deg1, also contains an amphipathic helix and exhibits 42% amino acid similarity with SM N100. Mutating SM residues Phe-35/Ser-37/Leu-65/Ile-69 into alanine, based on the key residues in Deg1, blunted SM cholesterol-mediated turnover. Taken together, our results support a model whereby the amphipathic helix in SM N100 attaches reversibly to the ER membrane depending on cholesterol levels; with excess, the helix is ejected and unravels, exposing a hydrophobic patch, which then serves as a degradation signal. Our findings shed new light on the regulation of a key cholesterol synthesis enzyme, highlighting the conservation of critical degron features from yeast to humans.
Keywords: cholesterol; cholesterol regulation; degron; endoplasmic reticulum (ER); membrane protein; protein degradation; squalene monooxygenase.
© 2017 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures



Comment in
-
A cholesterol-sensing mechanism unfolds.J Biol Chem. 2017 Dec 8;292(49):19974-19975. doi: 10.1074/jbc.H117.794230. J Biol Chem. 2017. PMID: 29222193 Free PMC article.
Similar articles
-
Non-canonical ubiquitination of the cholesterol-regulated degron of squalene monooxygenase.J Biol Chem. 2019 May 17;294(20):8134-8147. doi: 10.1074/jbc.RA119.007798. Epub 2019 Apr 2. J Biol Chem. 2019. PMID: 30940729 Free PMC article.
-
Valosin-containing protein mediates the ERAD of squalene monooxygenase and its cholesterol-responsive degron.Biochem J. 2019 Sep 20;476(18):2545-2560. doi: 10.1042/BCJ20190418. Biochem J. 2019. PMID: 31471528
-
The Degron Architecture of Squalene Monooxygenase and How Specific Lipids Calibrate Levels of This Key Cholesterol Synthesis Enzyme.Adv Exp Med Biol. 2021;21:1-12. doi: 10.1007/5584_2020_583. Adv Exp Med Biol. 2021. PMID: 32979157
-
Squalene monooxygenase: a journey to the heart of cholesterol synthesis.Prog Lipid Res. 2020 Jul;79:101033. doi: 10.1016/j.plipres.2020.101033. Epub 2020 Apr 28. Prog Lipid Res. 2020. PMID: 32360125 Review.
-
Post-translational control of the long and winding road to cholesterol.J Biol Chem. 2020 Dec 18;295(51):17549-17559. doi: 10.1074/jbc.REV120.010723. J Biol Chem. 2020. PMID: 33453997 Free PMC article. Review.
Cited by
-
Amphipathic helices sense the inner nuclear membrane environment through lipid packing defects.bioRxiv [Preprint]. 2024 Dec 20:2024.11.14.623600. doi: 10.1101/2024.11.14.623600. bioRxiv. 2024. PMID: 39605395 Free PMC article. Preprint.
-
Mechanism and therapeutic potential of liver injury induced by cholesterol-associated proteins.Front Pharmacol. 2025 Mar 27;16:1572592. doi: 10.3389/fphar.2025.1572592. eCollection 2025. Front Pharmacol. 2025. PMID: 40213681 Free PMC article. Review.
-
The shape of human squalene epoxidase expands the arsenal against cancer.Nat Commun. 2019 Feb 21;10(1):888. doi: 10.1038/s41467-019-08866-y. Nat Commun. 2019. PMID: 30792392 Free PMC article. Review.
-
Squalene and cholesterol in the balance at the ER membrane.Proc Natl Acad Sci U S A. 2020 Apr 14;117(15):8228-8230. doi: 10.1073/pnas.2003388117. Epub 2020 Apr 1. Proc Natl Acad Sci U S A. 2020. PMID: 32238557 Free PMC article. No abstract available.
-
An inducible amphipathic α-helix mediates subcellular targeting and membrane binding of RPE65.Life Sci Alliance. 2022 Oct 20;6(1):e202201546. doi: 10.26508/lsa.202201546. Print 2023 Jan. Life Sci Alliance. 2022. PMID: 36265895 Free PMC article.
References
-
- Gill S., Stevenson J., Kristiana I., and Brown A. J. (2011) Cholesterol-dependent degradation of squalene monooxygenase, a control point in cholesterol synthesis beyond HMG-CoA reductase. Cell Metab. 13, 260–273 - PubMed
-
- Ding J., Reynolds L. M., Zeller T., Müller C., Lohman K., Nicklas B. J., Kritchevsky S. B., Huang Z., de la Fuente A., Soranzo N., Settlage R. E., Chuang C.-C., Howard T., Xu N., Goodarzi M. O., et al. (2015) Alterations of a cellular cholesterol metabolism network are a molecular feature of obesity-related type 2 diabetes and cardiovascular disease. Diabetes 64, 3464–3474 - PMC - PubMed
-
- Buchovecky C. M., Turley S. D., Brown H. M., Kyle S. M., McDonald J. G., Liu B., Pieper A. A., Huang W., Katz D. M., Russell D. W., Shendure J., and Justice M. J. (2013) A suppressor screen in Mecp2 mutant mice implicates cholesterol metabolism in Rett syndrome. Nat. Genet. 45, 1013–1020 - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

