Activation and mechanism of a cryptic oviedomycin gene cluster via the disruption of a global regulatory gene, adpA, in Streptomyces ansochromogenes
- PMID: 28972184
- PMCID: PMC5712612
- DOI: 10.1074/jbc.M117.809145
Activation and mechanism of a cryptic oviedomycin gene cluster via the disruption of a global regulatory gene, adpA, in Streptomyces ansochromogenes
Abstract
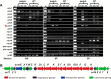
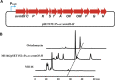
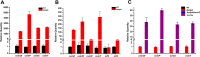
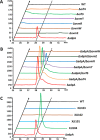
Genome sequencing analysis has revealed at least 35 clusters of likely biosynthetic genes for secondary metabolites in Streptomyces ansochromogenes. Disruption of adpA encoding a global regulator (AdpA) resulted in the failure of nikkomycin production, whereas other antibacterial activities against Staphylococcus aureus, Bacillus cereus, and Bacillus subtilis were observed with the fermentation broth of ΔadpA but not with that of the wild-type strain. Transcriptional analysis showed that a cryptic gene cluster (pks7), which shows high identity with an oviedomycin biosynthetic gene cluster (ovm), was activated in ΔadpA. The corresponding product of pks7 was characterized as oviedomycin by MS and NMR spectroscopy. To understand the molecular mechanism of ovm activation, the roles of six regulatory genes situated in the ovm cluster were investigated. Among them, proteins encoded by co-transcribed genes ovmZ and ovmW are positive regulators of ovm AdpA directly represses the transcription of ovmZ and ovmW Co-overexpression of ovmZ and ovmW can relieve the repression of AdpA on ovm transcription and effectively activate oviedomycin biosynthesis. The promoter of ovmOI-ovmH is identified as the direct target of OvmZ and OvmW. This is the first report that AdpA can simultaneously activate nikkomycin biosynthesis but repress oviedomycin biosynthesis in one strain. Our findings provide an effective strategy that is able to activate cryptic secondary metabolite gene clusters by genetic manipulation of global regulatory genes.
Keywords: AdpA; Streptomyces ansochromogenes; cryptic gene clusters; gene regulation; global regulators; metabolism; natural product biosynthesis; oviedomycin; polyketide; transcriptional coactivator.
© 2017 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures




References
-
- Baltz R. H. (2006) Marcel Faber Roundtable: is our antibiotic pipeline unproductive because of starvation, constipation or lack of inspiration? J. Ind. Microbiol. Biotechnol. 33, 507–513 - PubMed
-
- van Wezel G. P., McKenzie N. L., and Nodwell J. R. (2009) Chapter 5. Applying the genetics of secondary metabolism in model actinomycetes to the discovery of new antibiotics. Methods Enzymol. 458, 117–141 - PubMed
-
- Starcevic A., Zucko J., Simunkovic J., Long P. F., Cullum J., and Hranueli D. (2008) ClustScan: an integrated program package for the semi-automatic annotation of modular biosynthetic gene clusters and in silico prediction of novel chemical structures. Nucleic Acids Res. 36, 6882–6892 - PMC - PubMed
-
- Weber T., Rausch C., Lopez P., Hoof I., Gaykova V., Huson D. H., and Wohlleben W. (2009) CLUSEAN: a computer-based framework for the automated analysis of bacterial secondary metabolite biosynthetic gene clusters. J. Biotechnol. 140, 13–17 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

