Understanding DNA replication by the bacteriophage T4 replisome
- PMID: 28972188
- PMCID: PMC5682956
- DOI: 10.1074/jbc.R117.811208
Understanding DNA replication by the bacteriophage T4 replisome
Abstract
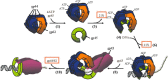
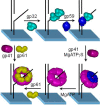
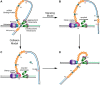
The T4 replisome has provided a unique opportunity to investigate the intricacies of DNA replication. We present a comprehensive review of this system focusing on the following: its 8-protein composition, their individual and synergistic activities, and assembly in vitro and in vivo into a replisome capable of coordinated leading/lagging strand DNA synthesis. We conclude with a brief comparison with other replisomes with emphasis on how coordinated DNA replication is achieved.
Keywords: DNA helicase; DNA polymerase; DNA primase; DNA replication; DNA-binding protein; bacteriophage.
© 2017 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures
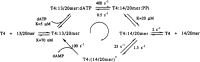
Similar articles
-
Binding Affinities among DNA Helicase-Primase, DNA Polymerase, and Replication Intermediates in the Replisome of Bacteriophage T7.J Biol Chem. 2016 Jan 15;291(3):1472-80. doi: 10.1074/jbc.M115.698233. Epub 2015 Nov 30. J Biol Chem. 2016. PMID: 26620561 Free PMC article.
-
Repetitive lagging strand DNA synthesis by the bacteriophage T4 replisome.Mol Biosyst. 2008 Nov;4(11):1070-4. doi: 10.1039/b812163j. Epub 2008 Sep 29. Mol Biosyst. 2008. PMID: 18931782 Review.
-
Interaction between the T4 helicase loading protein (gp59) and the DNA polymerase (gp43): unlocking of the gp59-gp43-DNA complex to initiate assembly of a fully functional replisome.Biochemistry. 2005 May 31;44(21):7747-56. doi: 10.1021/bi047296w. Biochemistry. 2005. PMID: 15909989
-
The Replication System of Bacteriophage T7.Enzymes. 2016;39:89-136. doi: 10.1016/bs.enz.2016.02.001. Epub 2016 Mar 28. Enzymes. 2016. PMID: 27241928 Review.
-
Dissociative properties of the proteins within the bacteriophage T4 replisome.J Biol Chem. 2003 Dec 12;278(50):49839-49. doi: 10.1074/jbc.M307405200. Epub 2003 Sep 18. J Biol Chem. 2003. PMID: 14500719
Cited by
-
Studies of DNA breathing in exciton-coupled (iCy3)2 dimer-labeled DNA constructs by polarization-sweep single-molecule fluorescence (PS-SMF) microscopy.Proc SPIE Int Soc Opt Eng. 2024 Jan-Feb;12863:128630C. doi: 10.1117/12.3001962. Epub 2024 Mar 13. Proc SPIE Int Soc Opt Eng. 2024. PMID: 39149417 Free PMC article.
-
Single-Molecule Insights Into the Dynamics of Replicative Helicases.Front Mol Biosci. 2021 Aug 26;8:741718. doi: 10.3389/fmolb.2021.741718. eCollection 2021. Front Mol Biosci. 2021. PMID: 34513934 Free PMC article. Review.
-
DNA Helicase-Polymerase Coupling in Bacteriophage DNA Replication.Viruses. 2021 Aug 31;13(9):1739. doi: 10.3390/v13091739. Viruses. 2021. PMID: 34578319 Free PMC article. Review.
-
Structural basis of the T4 bacteriophage primosome assembly and primer synthesis.Nat Commun. 2023 Jul 20;14(1):4396. doi: 10.1038/s41467-023-40106-2. Nat Commun. 2023. PMID: 37474605 Free PMC article.
-
Spectroscopic approaches for studies of site-specific DNA base and backbone 'breathing' using exciton-coupled dimer-labeled DNA.ArXiv [Preprint]. 2024 Mar 27:arXiv:2403.16251v2. ArXiv. 2024. PMID: 38584614 Free PMC article. Preprint.
References
-
- Mace D. C., and Alberts B. M. (1984) T4 DNA polymerase. Rates and processivity on single-stranded DNA templates. J. Mol. Biol. 177, 295–311 - PubMed
-
- Drake J. W. (1970) The Molecular Basis of Mutation, Holden-Day, San Francisco, CA
-
- Liu C. C., Burke R. L., Hibner U., Barry J., and Alberts B. (1979) Probing DNA replication mechanisms with the T4 bacteriophage in vitro system. Cold Spring Harb. Symp. Quant. Biol. 43, 469–487 - PubMed
-
- Alberts B. M., Barry J., Bedinger P., Formosa T., Jongeneel C. V., and Kreuzer K. N. (1983) Studies on DNA replication in the bacteriophage T4 in vitro system. Cold Spring Harb. Symp. Quant. Biol. 47, 655–668 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

