Quantitative proteomics reveals that long non-coding RNA MALAT1 interacts with DBC1 to regulate p53 acetylation
- PMID: 28973437
- PMCID: PMC5622371
- DOI: 10.1093/nar/gkx600
Quantitative proteomics reveals that long non-coding RNA MALAT1 interacts with DBC1 to regulate p53 acetylation
Erratum in
-
Corrigendum to article "Quantitative proteomics reveals that long non-coding RNA MALAT1 interacts with DBC1 to regulate p53 acetylation''.Nucleic Acids Res. 2021 Apr 19;49(7):4199-4202. doi: 10.1093/nar/gkab222. Nucleic Acids Res. 2021. PMID: 33751109 Free PMC article. No abstract available.
Abstract
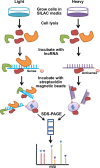
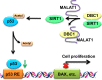
Metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) is a broadly expressed lncRNA involved in many aspects of cellular processes. To further delineate the underlying molecular mechanism, we employed a high-throughput strategy to characterize the interacting proteins of MALAT1 by combining RNA pull-down, quantitative proteomics, bioinformatics, and experimental validation. Our approach identified 127 potential MALAT1-interacting proteins and established a highly connected MALAT1 interactome network consisting of 788 connections. Gene ontology annotation and network analysis showed that MALAT1 was highly involved in five biological processes: RNA processing; gene transcription; ribosomal proteins; protein degradation; and metabolism regulation. The interaction between MALAT1 and depleted in breast cancer 1 (DBC1) was validated using RNA pull-down and RNA immunoprecipitation. Further mechanistic studies reveal that MALAT1 binding competes with the interaction between sirtuin1 (SIRT1) and DBC1, which then releases SIRT1 and enhances its deacetylation activity. Consequently, the deacetylation of p53 reduces the transcription of a spectrum of its downstream target genes, promotes cell proliferation and inhibits cell apoptosis. Our results uncover a novel mechanism by which MALAT1 regulates the activity of p53 through the lncRNA-protein interaction.
© The Author(s) 2017. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures




Comment in
-
Metastasis-associated lung adenocarcinoma transcript 1 regulates tumor progression: old wine in a new bottle.J Thorac Dis. 2018 Apr;10(Suppl 9):S1088-S1091. doi: 10.21037/jtd.2018.04.13. J Thorac Dis. 2018. PMID: 29850189 Free PMC article. No abstract available.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

