Direct modulation of T-box riboswitch-controlled transcription by protein synthesis inhibitors
- PMID: 28973457
- PMCID: PMC5622331
- DOI: 10.1093/nar/gkx663
Direct modulation of T-box riboswitch-controlled transcription by protein synthesis inhibitors
Abstract
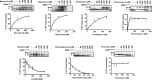
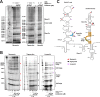
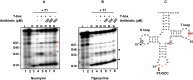
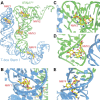
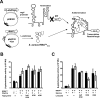
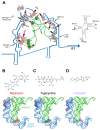
Recently, it was discovered that exposure to mainstream antibiotics activate numerous bacterial riboregulators that control antibiotic resistance genes including metabolite-binding riboswitches and other transcription attenuators. However, the effects of commonly used antibiotics, many of which exhibit RNA-binding properties, on the widespread T-box riboswitches, remain unknown. In Staphylococcus aureus, a species-specific glyS T-box controls the supply of glycine for both ribosomal translation and cell wall synthesis, making it a promising target for next-generation antimicrobials. Here, we report that specific protein synthesis inhibitors could either significantly increase T-box-mediated transcription antitermination, while other compounds could suppress it, both in vitro and in vivo. In-line probing of the full-length T-box combined with molecular modelling and docking analyses suggest that the antibiotics that promote transcription antitermination stabilize the T-box:tRNA complex through binding specific positions on stem I and the Staphylococcal-specific stem Sa. By contrast, the antibiotics that attenuate T-box transcription bind to other positions on stem I and do not interact with stem Sa. Taken together, our results reveal that the transcription of essential genes controlled by T-box riboswitches can be directly modulated by commonly used protein synthesis inhibitors. These findings accentuate the regulatory complexities of bacterial response to antimicrobials that involve multiple riboregulators.
© The Author(s) 2017. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

Similar articles
-
Structural idiosyncrasies of glycyl T-box riboswitches among pathogenic bacteria.RNA. 2024 Sep 16;30(10):1328-1344. doi: 10.1261/rna.080071.124. RNA. 2024. PMID: 38981655
-
Discovery of Small-Molecule Antibiotics against a Unique tRNA-Mediated Regulation of Transcription in Gram-Positive Bacteria.ChemMedChem. 2019 Apr 3;14(7):758-769. doi: 10.1002/cmdc.201800744. Epub 2019 Mar 1. ChemMedChem. 2019. PMID: 30707489
-
Lineage-specific insertions in T-box riboswitches modulate antibiotic binding and action.Nucleic Acids Res. 2022 Jun 10;50(10):5834-5849. doi: 10.1093/nar/gkac359. Nucleic Acids Res. 2022. PMID: 35580054 Free PMC article.
-
An evolving tale of two interacting RNAs-themes and variations of the T-box riboswitch mechanism.IUBMB Life. 2019 Aug;71(8):1167-1180. doi: 10.1002/iub.2098. Epub 2019 Jun 17. IUBMB Life. 2019. PMID: 31206978 Free PMC article. Review.
-
Riboswitches: discovery of drugs that target bacterial gene-regulatory RNAs.Acc Chem Res. 2011 Dec 20;44(12):1329-38. doi: 10.1021/ar200039b. Epub 2011 May 26. Acc Chem Res. 2011. PMID: 21615107 Free PMC article. Review.
Cited by
-
Ribocentre-switch: a database of riboswitches.Nucleic Acids Res. 2024 Jan 5;52(D1):D265-D272. doi: 10.1093/nar/gkad891. Nucleic Acids Res. 2024. PMID: 37855663 Free PMC article.
-
The T-Box Riboswitch: tRNA as an Effector to Modulate Gene Regulation.Microbiol Spectr. 2018 Jul;6(4):10.1128/microbiolspec.rwr-0028-2018. doi: 10.1128/microbiolspec.RWR-0028-2018. Microbiol Spectr. 2018. PMID: 30051797 Free PMC article. Review.
-
Another layer of complexity in Staphylococcus aureus methionine biosynthesis control: unusual RNase III-driven T-box riboswitch cleavage determines met operon mRNA stability and decay.Nucleic Acids Res. 2021 Feb 26;49(4):2192-2212. doi: 10.1093/nar/gkaa1277. Nucleic Acids Res. 2021. PMID: 33450025 Free PMC article.
-
Unboxing the T-box riboswitches-A glimpse into multivalent and multimodal RNA-RNA interactions.Wiley Interdiscip Rev RNA. 2020 Nov;11(6):e1600. doi: 10.1002/wrna.1600. Epub 2020 Jul 6. Wiley Interdiscip Rev RNA. 2020. PMID: 32633085 Free PMC article. Review.
-
A Riboswitch-Driven Era of New Antibacterials.Antibiotics (Basel). 2022 Sep 13;11(9):1243. doi: 10.3390/antibiotics11091243. Antibiotics (Basel). 2022. PMID: 36140022 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

