Direct modulation of T-box riboswitch-controlled transcription by protein synthesis inhibitors
- PMID: 28973457
- PMCID: PMC5622331
- DOI: 10.1093/nar/gkx663
Direct modulation of T-box riboswitch-controlled transcription by protein synthesis inhibitors
Abstract
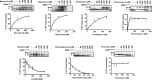
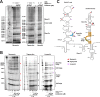
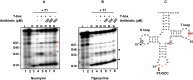
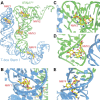
Recently, it was discovered that exposure to mainstream antibiotics activate numerous bacterial riboregulators that control antibiotic resistance genes including metabolite-binding riboswitches and other transcription attenuators. However, the effects of commonly used antibiotics, many of which exhibit RNA-binding properties, on the widespread T-box riboswitches, remain unknown. In Staphylococcus aureus, a species-specific glyS T-box controls the supply of glycine for both ribosomal translation and cell wall synthesis, making it a promising target for next-generation antimicrobials. Here, we report that specific protein synthesis inhibitors could either significantly increase T-box-mediated transcription antitermination, while other compounds could suppress it, both in vitro and in vivo. In-line probing of the full-length T-box combined with molecular modelling and docking analyses suggest that the antibiotics that promote transcription antitermination stabilize the T-box:tRNA complex through binding specific positions on stem I and the Staphylococcal-specific stem Sa. By contrast, the antibiotics that attenuate T-box transcription bind to other positions on stem I and do not interact with stem Sa. Taken together, our results reveal that the transcription of essential genes controlled by T-box riboswitches can be directly modulated by commonly used protein synthesis inhibitors. These findings accentuate the regulatory complexities of bacterial response to antimicrobials that involve multiple riboregulators.
© The Author(s) 2017. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

