MeCP2_E1 N-terminal modifications affect its degradation rate and are disrupted by the Ala2Val Rett mutation
- PMID: 28973632
- PMCID: PMC5886153
- DOI: 10.1093/hmg/ddx300
MeCP2_E1 N-terminal modifications affect its degradation rate and are disrupted by the Ala2Val Rett mutation
Abstract
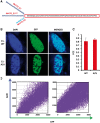
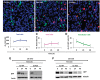
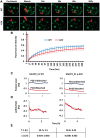
Methyl CpG-binding protein 2 (MeCP2), the mutated protein in Rett syndrome (RTT), is a crucial chromatin-modifying and gene-regulatory protein that has two main isoforms (MeCP2_E1 and MeCP2_ E2) due to the alternative splicing and switching between translation start codons in exons one and two. Functionally, these two isoforms appear to be virtually identical; however, evidence suggests that only MeCP2_E1 is relevant to RTT, including a single RTT missense mutation in exon 1, Ala2Val. Here, we show that N-terminal co- and post-translational modifications differ for MeCP2_E1 and MeCP2_E1-Ala2Val, which result in different protein degradation rates in vitro. We report complete N-methionine excision (NME) for MeCP2_E1 and evidence of excision of multiple alanine residues from the N-terminal polyalanine stretch. For MeCP2_E1-Ala2Val, we observed only partial NME and N-acetylation (NA) of either methionine or valine. The localization of MeCP2_E1 and co-localization with chromatin appear to be unaffected by the Ala2Val mutation. However, a higher proteasomal degradation rate was observed for MeCP2_E1-Ala2Val compared with that for wild type MeCP2_E1. Thus, the etiopathology of Ala2Val is likely due to a reduced bio-availability of MeCP2 because of the faster degradation rate of the unmodified defective protein. Our data on the effects of the Ala2Val mutation on N-terminal modifications of MeCP2 may be applicable to Ala2Val mutations in other disease genes for which no etiopathological mechanism has been established.
© The Author 2017. Published by Oxford University Press. All rights reserved. For Permissions, please email: journals.permissions@oup.com.
Figures

References
-
- Lewis J.D., Meehan R.R., Henzel W.J., Maurer-Fogy I., Jeppesen P., Klein F., Bird A. (1992) Purification, sequence, and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell, 69, 905–914. - PubMed
-
- Amir R.E., Van den Veyver I.B., Wan M., Tran C.Q., Francke U., Zoghbi H.Y. (1999) Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat. Genet., 23, 185–188. - PubMed
-
- Reichwald K., Thiesen J., Wiehe T., Weitzel J., Strätling W.H., Kioschis P., Poustka A., Rosenthal A., Platzer M. (2000) Comparative sequence analysis of the MECP2-locus in human and mouse reveals new transcribed regions. Mamm. Genome, 11, 182–190. - PubMed
-
- Mnatzakanian G.N., Lohi H., Munteanu I., Alfred S.E., Yamada T., MacLeod P.J.M., Jones J.R., Scherer S.W., Schanen N.C., Friez M.J., Vincent J.B., Minassian B.A. (2004) A previously unidentified MECP2 open reading frame defines a new protein isoform relevant to Rett syndrome. Nat. Genet., 36, 339–341. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

