Alpha-synuclein induces the unfolded protein response in Parkinson's disease SNCA triplication iPSC-derived neurons
- PMID: 28973645
- PMCID: PMC5886237
- DOI: 10.1093/hmg/ddx331
Alpha-synuclein induces the unfolded protein response in Parkinson's disease SNCA triplication iPSC-derived neurons
Abstract
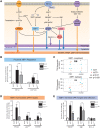
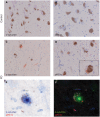
The recent generation of induced pluripotent stem cells (iPSCs) from a patient with Parkinson's disease (PD) resulting from triplication of the α-synuclein (SNCA) gene locus allows unprecedented opportunities to explore its contribution to the molecular pathogenesis of PD. We used the double-nicking CRISPR/Cas9 system to conduct site-specific mutagenesis of SNCA in these cells, generating an isogenic iPSC line with normalized SNCA gene dosage. Comparative gene expression analysis of neuronal derivatives from these iPSCs revealed an ER stress phenotype, marked by induction of the IRE1α/XBP1 axis of the unfolded protein response (UPR) and culminating in terminal UPR activation. Neuropathological analysis of post-mortem brain tissue demonstrated that pIRE1α is expressed in PD brains within neurons containing elevated levels of α-synuclein or Lewy bodies. Having used this pair of isogenic iPSCs to define this phenotype, these cells can be further applied in UPR-targeted drug discovery towards the development of disease-modifying therapeutics.
© The Author 2017. Published by Oxford University Press. All rights reserved. For Permissions, please email: journals.permissions@oup.com.
Figures


References
-
- Singleton A.B., Farrer M., Johnson J., Singleton A., Hague S., Kachergus J., Hulihan M., Peuralinna T., Dutra A., Nussbaum R. (2003) Alpha-synuclein locus triplication causes Parkinson's disease. Science, 302, 841.. - PubMed
-
- Chartier-Harlin M.C., Kachergus J., Roumier C., Mouroux V., Douay X., Lincoln S., Levecque C., Larvor L., Andrieux J., Hulihan M.. et al. (2004) Alpha-synuclein locus duplication as a cause of familial Parkinson's disease. Lancet, 364, 1167–1169. - PubMed
-
- Ibanez P., Bonnet A.M., Debarges B., Lohmann E., Tison F., Pollak P., Agid Y., Durr A., Brice A. (2004) Causal relation between alpha-synuclein gene duplication and familial Parkinson's disease. Lancet, 364, 1169–1171. - PubMed
-
- Farrer M., Kachergus J., Forno L., Lincoln S., Wang D.S., Hulihan M., Maraganore D., Gwinn-Hardy K., Wszolek Z., Dickson D.. et al. (2004) Comparison of kindreds with parkinsonism and alpha-synuclein genomic multiplications. Ann. Neurol., 55, 174–179. - PubMed
-
- Singleton A.C. J. (2007) Dawson T. (ed.), In Parkinson's Disease: Genetics and Pathogenesis. Informa Healthcare USA, Inc, New York, NY, in press., pp. 19–33.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

