Peanuts that keep aflatoxin at bay: a threshold that matters
- PMID: 28973784
- PMCID: PMC5902767
- DOI: 10.1111/pbi.12846
Peanuts that keep aflatoxin at bay: a threshold that matters
Abstract
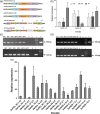
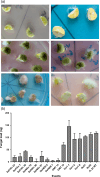
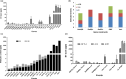
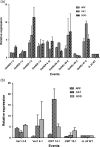
Aflatoxin contamination in peanuts poses major challenges for vulnerable populations of sub-Saharan Africa and South Asia. Developing peanut varieties to combat preharvest Aspergillus flavus infection and resulting aflatoxin contamination has thus far remained a major challenge, confounded by highly complex peanut-Aspergilli pathosystem. Our study reports achieving a high level of resistance in peanut by overexpressing (OE) antifungal plant defensins MsDef1 and MtDef4.2, and through host-induced gene silencing (HIGS) of aflM and aflP genes from the aflatoxin biosynthetic pathway. While the former improves genetic resistance to A. flavus infection, the latter inhibits aflatoxin production in the event of infection providing durable resistance against different Aspergillus flavus morphotypes and negligible aflatoxin content in several peanut events/lines well. A strong positive correlation was observed between aflatoxin accumulation and decline in transcription of the aflatoxin biosynthetic pathway genes in both OE-Def and HIGS lines. Transcriptomic signatures in the resistant lines revealed key mechanisms such as regulation of aflatoxin synthesis, its packaging and export control, besides the role of reactive oxygen species-scavenging enzymes that render enhanced protection in the OE and HIGS lines. This is the first study to demonstrate highly effective biotechnological strategies for successfully generating peanuts that are near-immune to aflatoxin contamination, offering a panacea for serious food safety, health and trade issues in the semi-arid regions.
Keywords: Aspergillus flavus; aflatoxin; defensins; food safety; host-induced gene silencing; mycotoxins; peanut.
© 2017 The Authors. Plant Biotechnology Journal published by Society for Experimental Biology and The Association of Applied Biologists and John Wiley & Sons Ltd.
Figures

References
-
- Alkhayyat, F. and Yu, J.H. (2014) Upstream regulation of mycotoxin biosynthesis. Adv. Appl. Microbiol. 86, 251–278. - PubMed
-
- Bhatnagar, D. , Ehrlich, K.C. and Cleveland, T.E. (1991) Oxidation‐reduction reactions in biosynthesis of secondary metabolites. In Handbook of Applied Mycology: mycotoxins in ecological systems, vol. 5 ( Bhatnagar, D. , Lillehoj, E.B. and Arora, D.K. , eds), pp. 255–286. New York: Marcel Dekker Inc.
-
- Bhatnagar, M. , Prasad, K. , Bhatnagar‐Mathur, P. , Narasu, M.L. , Waliyar, F. and Sharma, K.K. (2010) An efficient method for the production of marker‐free transgenic plants of peanut (Arachis hypogaea L.). Plant Cell Rep. 29, 495–502. - PubMed
-
- Bhatnagar‐Mathur, P. , Sunkara, S. , Bhatnagar‐Panwar, M. , Waliyar, F. and Sharma, K.K. (2015) Biotechnological advances for combating Aspergillus flavus and aflatoxin contamination in crops. Plant Sci. 234, 119–132. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

