Structural basis underlying complex assembly and conformational transition of the type I R-M system
- PMID: 28973912
- PMCID: PMC5651777
- DOI: 10.1073/pnas.1711754114
Structural basis underlying complex assembly and conformational transition of the type I R-M system
Abstract
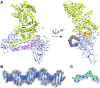
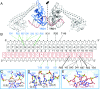
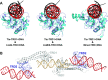
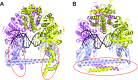
Type I restriction-modification (R-M) systems are multisubunit enzymes with separate DNA-recognition (S), methylation (M), and restriction (R) subunits. Despite extensive studies spanning five decades, the detailed molecular mechanisms underlying subunit assembly and conformational transition are still unclear due to the lack of high-resolution structural information. Here, we report the atomic structure of a type I MTase complex (2M+1S) bound to DNA and cofactor S-adenosyl methionine in the "open" form. The intermolecular interactions between M and S subunits are mediated by a four-helix bundle motif, which also determines the specificity of the interaction. Structural comparison between open and previously reported low-resolution "closed" structures identifies the huge conformational changes within the MTase complex. Furthermore, biochemical results show that R subunits prefer to load onto the closed form MTase. Based on our results, we proposed an updated model for the complex assembly. The work reported here provides guidelines for future applications in molecular biology.
Keywords: EcoKI; MTase; crystal structure; type I R-M system.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






Comment in
-
Structures of the type I DNA restriction enzymes.Proc Natl Acad Sci U S A. 2017 Nov 28;114(48):E10261-E10262. doi: 10.1073/pnas.1718623114. Epub 2017 Nov 14. Proc Natl Acad Sci U S A. 2017. PMID: 29138322 Free PMC article. No abstract available.
Similar articles
-
Structural insights into assembly, operation and inhibition of a type I restriction-modification system.Nat Microbiol. 2020 Sep;5(9):1107-1118. doi: 10.1038/s41564-020-0731-z. Epub 2020 Jun 1. Nat Microbiol. 2020. PMID: 32483229
-
Structure of HsdS subunit from Thermoanaerobacter tengcongensis sheds lights on mechanism of dynamic opening and closing of type I methyltransferase.PLoS One. 2011 Mar 2;6(3):e17346. doi: 10.1371/journal.pone.0017346. PLoS One. 2011. PMID: 21399684 Free PMC article.
-
Structural features of a minimal intact methyltransferase of a type I restriction-modification system.Int J Biol Macromol. 2022 May 31;208:381-389. doi: 10.1016/j.ijbiomac.2022.03.115. Epub 2022 Mar 23. Int J Biol Macromol. 2022. PMID: 35337914
-
Novel m4C modification in type I restriction-modification systems.Nucleic Acids Res. 2016 Nov 2;44(19):9413-9425. doi: 10.1093/nar/gkw743. Epub 2016 Aug 31. Nucleic Acids Res. 2016. PMID: 27580720 Free PMC article.
-
Role of Restriction-Modification Systems in Prokaryotic Evolution and Ecology.Biochemistry (Mosc). 2015 Oct;80(10):1373-86. doi: 10.1134/S0006297915100193. Biochemistry (Mosc). 2015. PMID: 26567582 Review.
Cited by
-
Structural insights into assembly, operation and inhibition of a type I restriction-modification system.Nat Microbiol. 2020 Sep;5(9):1107-1118. doi: 10.1038/s41564-020-0731-z. Epub 2020 Jun 1. Nat Microbiol. 2020. PMID: 32483229
-
Cryo-EM structures of DNA-free and DNA-bound BsaXI: architecture of a Type IIB restriction-modification enzyme.Nucleic Acids Res. 2025 Apr 10;53(7):gkaf291. doi: 10.1093/nar/gkaf291. Nucleic Acids Res. 2025. PMID: 40239994 Free PMC article.
-
Coordination of phage genome degradation versus host genome protection by a bifunctional restriction-modification enzyme visualized by CryoEM.Structure. 2021 Jun 3;29(6):521-530.e5. doi: 10.1016/j.str.2021.03.012. Epub 2021 Apr 6. Structure. 2021. PMID: 33826880 Free PMC article.
-
Bacterial DNA methyltransferase: A key to the epigenetic world with lessons learned from proteobacteria.Front Microbiol. 2023 Mar 22;14:1129437. doi: 10.3389/fmicb.2023.1129437. eCollection 2023. Front Microbiol. 2023. PMID: 37032876 Free PMC article. Review.
-
A model for the evolution of prokaryotic DNA restriction-modification systems based upon the structural malleability of Type I restriction-modification enzymes.Nucleic Acids Res. 2018 Sep 28;46(17):9067-9080. doi: 10.1093/nar/gky760. Nucleic Acids Res. 2018. PMID: 30165537 Free PMC article.
References
-
- Dupuis ME, Villion M, Magadán AH, Moineau S. CRISPR-Cas and restriction-modification systems are compatible and increase phage resistance. Nat Commun. 2013;4:2087. - PubMed
-
- Wright AV, Nuñez JK, Doudna JA. Biology and applications of CRISPR systems: Harnessing nature’s toolbox for genome engineering. Cell. 2016;164:29–44. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

