Mediator subunit MED25 links the jasmonate receptor to transcriptionally active chromatin
- PMID: 28973940
- PMCID: PMC5651773
- DOI: 10.1073/pnas.1710885114
Mediator subunit MED25 links the jasmonate receptor to transcriptionally active chromatin
Abstract
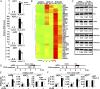
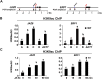
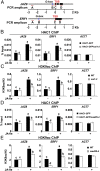
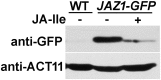
Jasmonoyl-isoleucine (JA-Ile), the active form of the plant hormone jasmonate (JA), is sensed by the F-box protein CORONATINE INSENSITIVE 1 (COI1), a component of a functional Skp-Cullin-F-box E3 ubiquitin ligase complex. Sensing of JA-Ile by COI1 rapidly triggers genome-wide transcriptional changes that are largely regulated by the basic helix-loop-helix transcription factor MYC2. However, it remains unclear how the JA-Ile receptor protein COI1 relays hormone-specific regulatory signals to the RNA polymerase II general transcriptional machinery. Here, we report that the plant transcriptional coactivator complex Mediator directly links COI1 to the promoters of MYC2 target genes. MED25, a subunit of the Mediator complex, brings COI1 to MYC2 target promoters and facilitates COI1-dependent degradation of jasmonate-ZIM domain (JAZ) transcriptional repressors. MED25 and COI1 influence each other's enrichment on MYC2 target promoters. Furthermore, MED25 physically and functionally interacts with HISTONE ACETYLTRANSFERASE1 (HAC1), which plays an important role in JA signaling by selectively regulating histone (H) 3 lysine (K) 9 (H3K9) acetylation of MYC2 target promoters. Moreover, the enrichment and function of HAC1 on MYC2 target promoters depend on COI1 and MED25. Therefore, the MED25 interface of Mediator links COI1 with HAC1-dependent H3K9 acetylation to activate MYC2-regulated transcription of JA-responsive genes. This study exemplifies how a single Mediator subunit integrates the actions of both genetic and epigenetic regulators into a concerted transcriptional program.
Keywords: COI1; MED25; MYC2; jasmonate; nuclear hormone receptor.
Conflict of interest statement
The authors declare no conflict of interest.
Figures








References
-
- Browse J. Jasmonate passes muster: A receptor and targets for the defense hormone. Annu Rev Plant Biol. 2009;60:183–205. - PubMed
-
- Xie DX, Feys BF, James S, Nieto-Rostro M, Turner JG. COI1: An Arabidopsis gene required for jasmonate-regulated defense and fertility. Science. 1998;280:1091–1094. - PubMed
-
- Thines B, et al. JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature. 2007;448:661–665. - PubMed
-
- Chini A, et al. The JAZ family of repressors is the missing link in jasmonate signalling. Nature. 2007;448:666–671. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

