Species and genotype diversity of Plasmodium in malaria patients from Gabon analysed by next generation sequencing
- PMID: 28974215
- PMCID: PMC5627438
- DOI: 10.1186/s12936-017-2044-0
Species and genotype diversity of Plasmodium in malaria patients from Gabon analysed by next generation sequencing
Abstract
Background: Six Plasmodium species are known to naturally infect humans. Mixed species infections occur regularly but morphological discrimination by microscopy is difficult and multiplicity of infection (MOI) can only be evaluated by molecular methods. This study investigated the complexity of Plasmodium infections in patients treated for microscopically detected non-falciparum or mixed species malaria in Gabon.
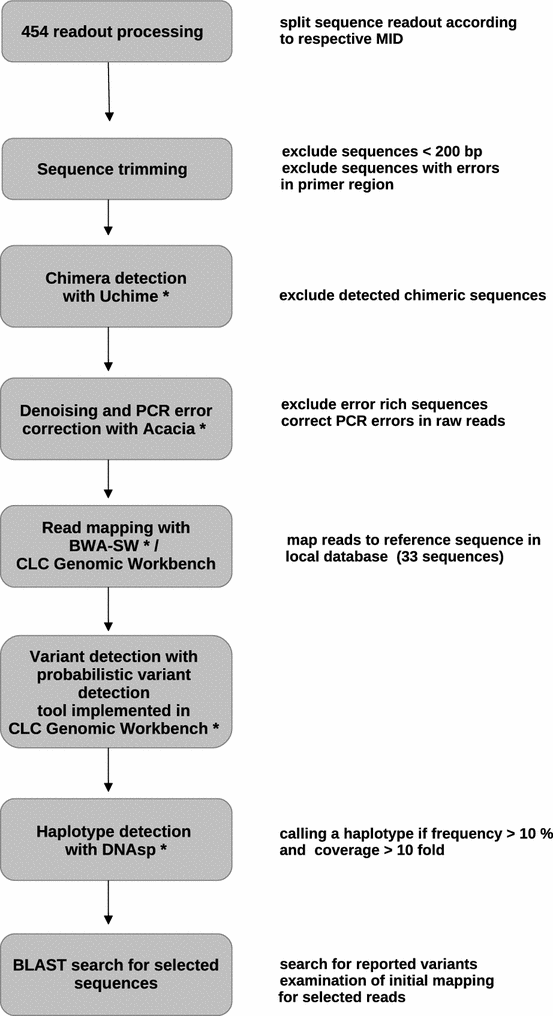
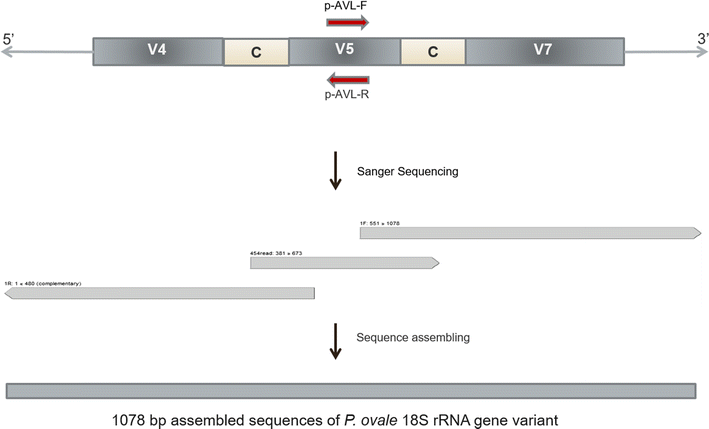
Methods: Ultra-deep sequencing of nucleus (18S rRNA), mitochondrion, and apicoplast encoded genes was used to evaluate Plasmodium species diversity and MOI in 46 symptomatic Gabonese patients with microscopically diagnosed non-falciparum or mixed species malaria.
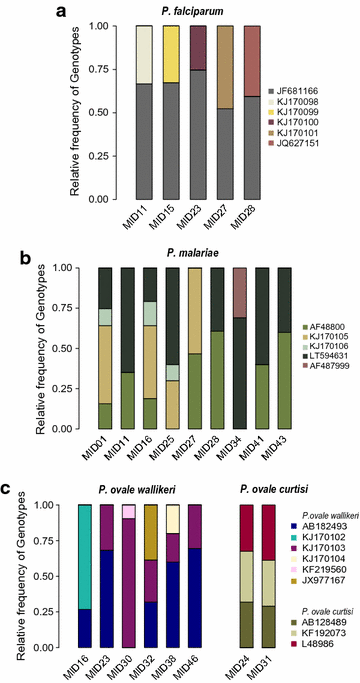
Results: Deep sequencing revealed a large complexity of confections in patients with uncomplicated malaria, both on species and genotype levels. Mixed infections involved up to four parasite species (Plasmodium falciparum, Plasmodium malariae, Plasmodium ovale curtisi, and P. ovale wallikeri). Multiple genotypes from each species were determined from the asexual 18S rRNA gene. 17 of 46 samples (37%) harboured multiple genotypes of at least one Plasmodium species. The number of genotypes per sample (MOI) was highest in P. malariae (n = 4), followed by P. ovale curtisi (n = 3), P. ovale wallikeri (n = 3), and P. falciparum (n = 2). The highest combined genotype complexity in samples that contained mixed-species infections was seven.
Conclusions: Ultra-deep sequencing showed an unexpected breadth of Plasmodium species and within species diversity in clinical samples. MOI of P. ovale curtisi, P. ovale wallikeri and P. malariae infections were higher than anticipated and contribute significantly to the burden of malaria in Gabon.
Keywords: Amplicon; Metagenomics; Next-generation sequencing; Plasmodium falciparum; Plasmodium malariae; Plasmodium ovale curtisi; Plasmodium ovale wallikeri.
Figures
References
-
- Ansari HR, Templeton TJ, Subudhi AK, Ramaprasad A, Tang J, Lu F, et al. Genome-scale comparison of expanded gene families in Plasmodium ovale wallikeri and Plasmodium ovale curtisi with Plasmodium malariae and with other Plasmodium species. Int J Parasitol. 2016;46:685–696. doi: 10.1016/j.ijpara.2016.05.009. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

