The stem cell factor SALL4 is an essential transcriptional regulator in mixed lineage leukemia-rearranged leukemogenesis
- PMID: 28974232
- PMCID: PMC5627455
- DOI: 10.1186/s13045-017-0531-y
The stem cell factor SALL4 is an essential transcriptional regulator in mixed lineage leukemia-rearranged leukemogenesis
Abstract
Background: The stem cell factor spalt-like transcription factor 4 (SALL4) plays important roles in normal hematopoiesis and also in leukemogenesis. We previously reported that SALL4 exerts its effect by recruiting important epigenetic factors such as DNA methyltransferases DNMT1 and lysine-specific demethylase 1 (LSD1/KDM1A). Both of these proteins are critically involved in mixed lineage leukemia (MLL)-rearranged (MLL-r) leukemia, which has a very poor clinical prognosis. Recently, SALL4 has been further linked to the functions of MLL and its target gene homeobox A9 (HOXA9). However, it remains unclear whether SALL4 is indeed a key player in MLL-r leukemia pathogenesis.
Methods: Using a mouse bone marrow retroviral transduction/ transplantation approach combined with tamoxifen-inducible, CreERT2-mediated Sall4 gene deletion, we studied SALL4 functions in leukemic transformation that was induced by MLL-AF9-one of the most common MLL-r oncoproteins found in patients. In addition, the underlying transcriptional and epigenetic mechanisms were explored using chromatin immunoprecipitation (ChIP) sequencing (ChIP-Seq), mRNA microarray, qRT-PCR, histone modification, co-immunoprecipitation (co-IP), cell cycle, and apoptosis assays. The effects of SALL4 loss on normal hematopoiesis in mice were also investigated.
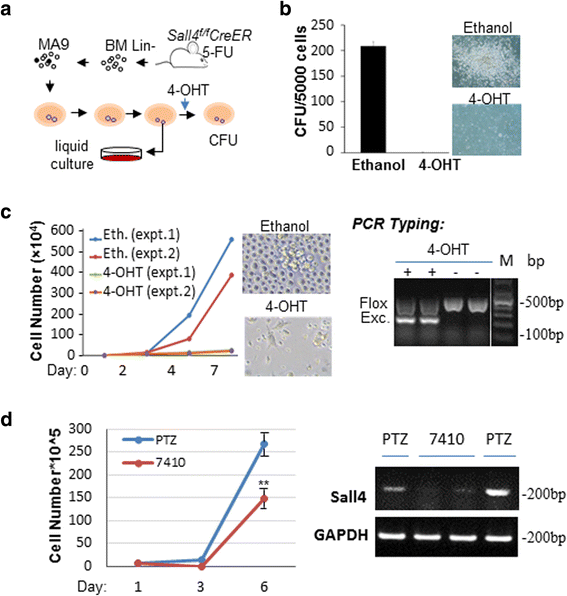
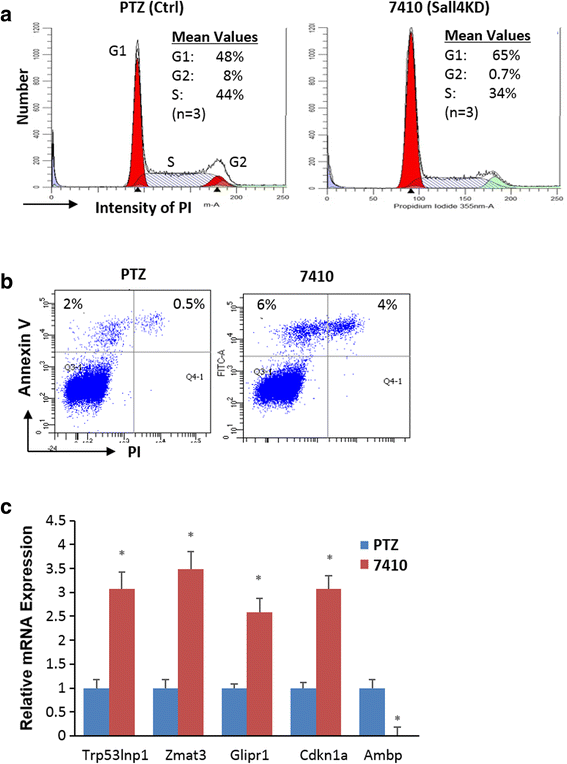
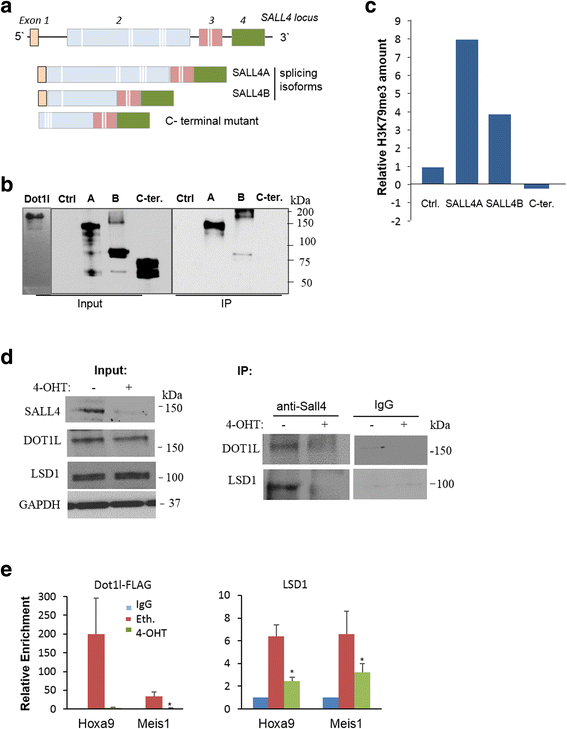
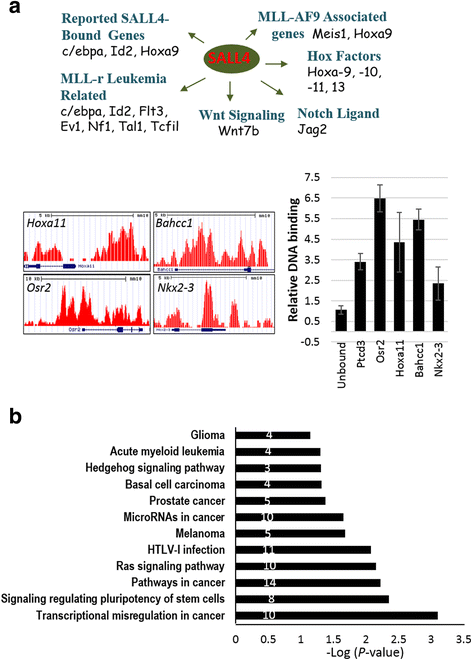
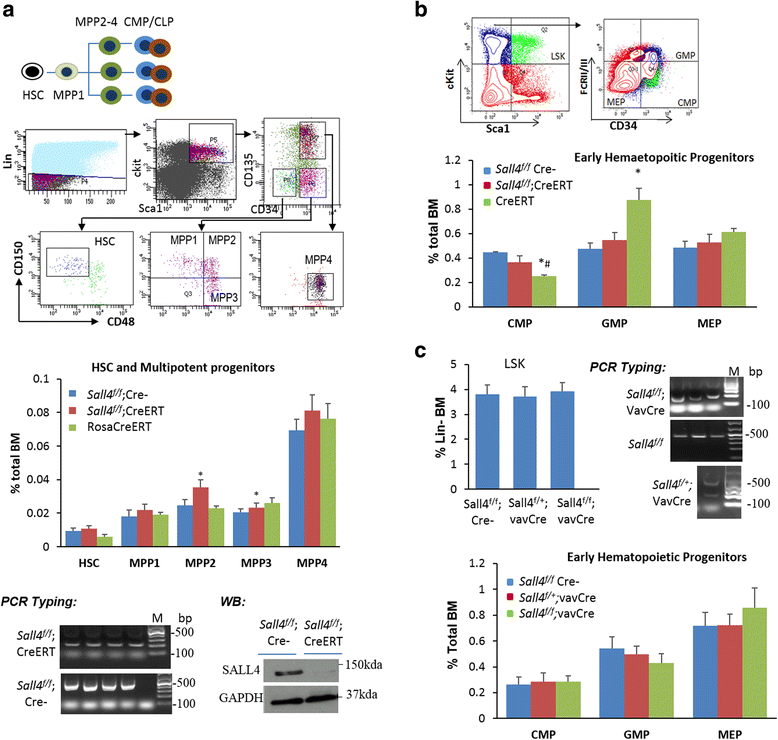
Results: In vitro and in vivo studies revealed that SALL4 expression is critically required for MLL-AF9-induced leukemic transformation and disease progression in mice. Loss of SALL4 in MLL-AF9-transformed cells induced apoptosis and cell cycle arrest at G1. ChIP-Seq assay identified that Sall4 binds to key MLL-AF9 target genes and important MLL-r or non-MLL-r leukemia-related genes. ChIP-PCR assays indicated that SALL4 affects the levels of the histone modification markers H3K79me2/3 and H3K4me3 at MLL-AF9 target gene promoters by physically interacting with DOT1-like histone H3K79 methyltransferase (DOT1l) and LSD1/KDM1A, and thereby regulates transcript expression. Surprisingly, normal Sall4 f/f /CreERT2 mice treated with tamoxifen or vav-Cre-mediated (hematopoietic-specific) Sall4 -/- mice were healthy and displayed no significant hematopoietic defects.
Conclusions: Our findings indicate that SALL4 critically contributes to MLL-AF9-induced leukemia, unraveling the underlying transcriptional and epigenetic mechanisms in this disease and suggesting that selectively targeting the SALL4 pathway may be a promising approach for managing human MLL-r leukemia.
Keywords: DOT1l; Epigenetic; Hematopoietic stem cells; Histone methylation; LSD1; Transcription factor.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures


References
-
- Kohlhase J, Schuh R, Dowe G, Kuhnlein RP, Jackle H, Schroeder B, Schulz-Schaeffer W, Kretzschmar HA, Kohler A, Muller U, et al. Isolation, characterization, and organ-specific expression of two novel human zinc finger genes related to the Drosophila gene spalt. Genomics. 1996;38(3):291–298. doi: 10.1006/geno.1996.0631. - DOI - PubMed
-
- Al-Baradie R, Yamada K, St Hilaire C, Chan WM, Andrews C, McIntosh N, Nakano M, Martonyi EJ, Raymond WR, Okumura S, et al. Duane radial ray syndrome (Okihiro syndrome) maps to 20q13 and results from mutations in SALL4, a new member of the SAL family. Am J Hum Genet. 2002;71(5):1195–1199. doi: 10.1086/343821. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

