Preparing for the crewed Mars journey: microbiota dynamics in the confined Mars500 habitat during simulated Mars flight and landing
- PMID: 28974259
- PMCID: PMC5627443
- DOI: 10.1186/s40168-017-0345-8
Preparing for the crewed Mars journey: microbiota dynamics in the confined Mars500 habitat during simulated Mars flight and landing
Abstract
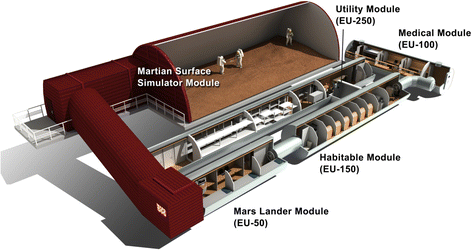
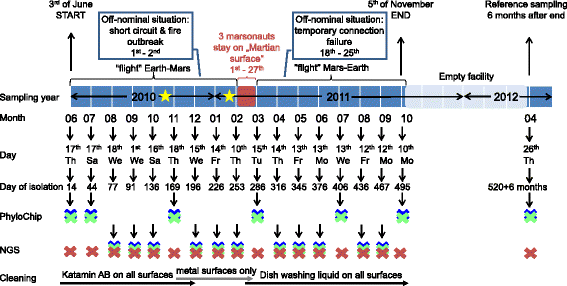
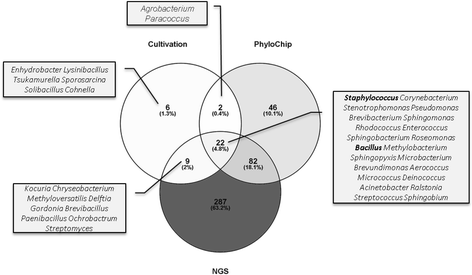
Background: The Mars500 project was conceived as the first full duration simulation of a crewed return flight to Mars. For 520 days, six crew members lived confined in a specifically designed spacecraft mock-up. The herein described "MIcrobial ecology of Confined Habitats and humAn health" (MICHA) experiment was implemented to acquire comprehensive microbiota data from this unique, confined manned habitat, to retrieve important information on the occurring microbiota dynamics, the microbial load and diversity in the air and on various surfaces. In total, 360 samples from 20 (9 air, 11 surface) locations were taken at 18 time-points and processed by extensive cultivation, PhyloChip and next generation sequencing (NGS) of 16S rRNA gene amplicons.
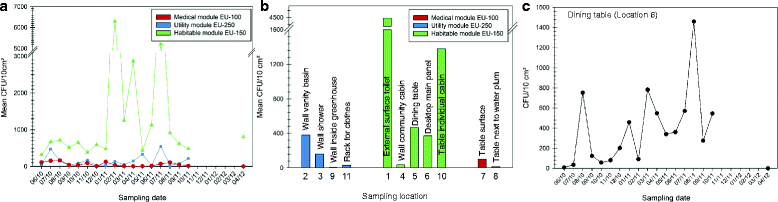
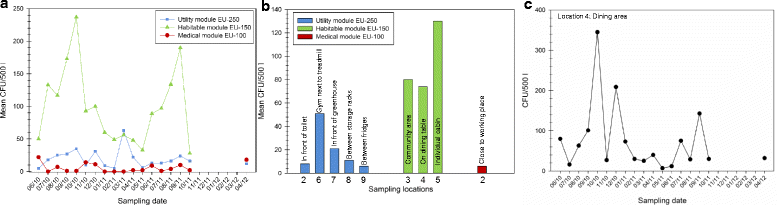
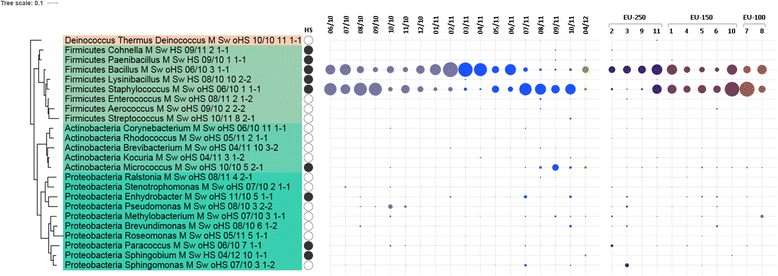
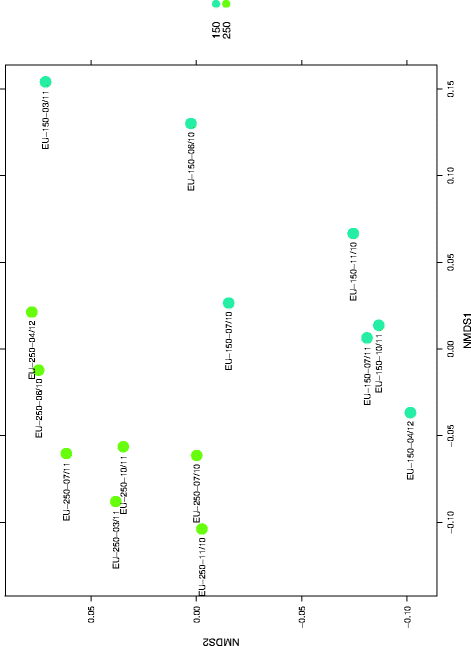
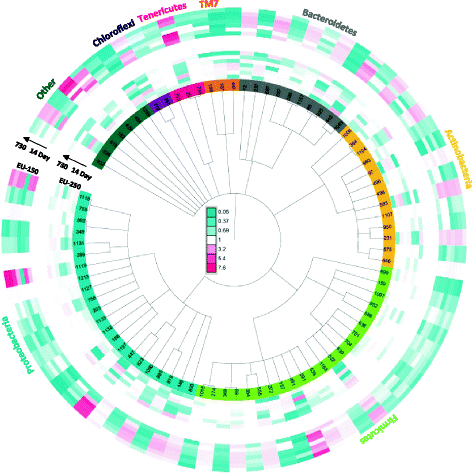
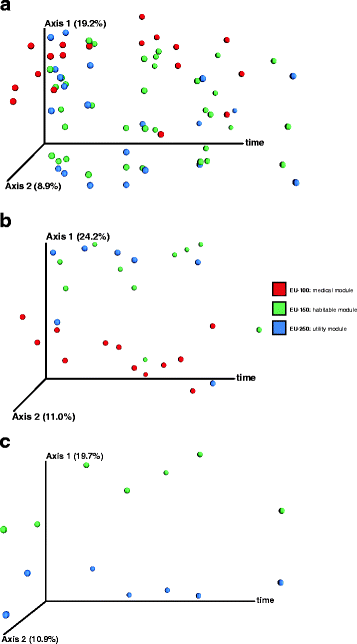
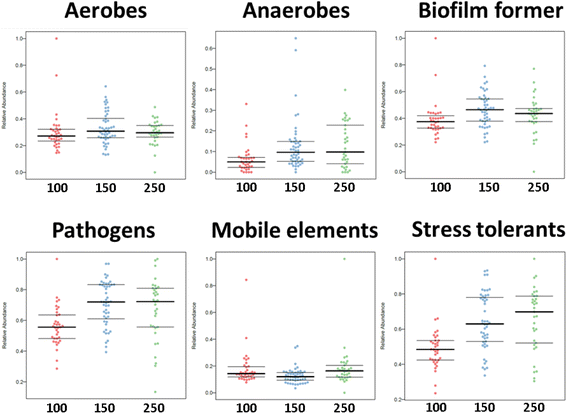
Results: Cultivation assays revealed a Staphylococcus and Bacillus-dominated microbial community on various surfaces, with an average microbial load that did not exceed the allowed limits for ISS in-flight requirements indicating adequate maintenance of the facility. Areas with high human activity were identified as hotspots for microbial accumulation. Despite substantial fluctuation with respect to microbial diversity and abundance throughout the experiment, the location within the facility and the confinement duration were identified as factors significantly shaping the microbial diversity and composition, with the crew representing the main source for microbial dispersal. Opportunistic pathogens, stress-tolerant or potentially mobile element-bearing microorganisms were predicted to be prevalent throughout the confinement, while the overall microbial diversity dropped significantly over time.
Conclusions: Our findings clearly indicate that under confined conditions, the community structure remains a highly dynamic system which adapts to the prevailing habitat and micro-conditions. Since a sterile environment is not achievable, these dynamics need to be monitored to avoid spreading of highly resistant or potentially pathogenic microorganisms and a potentially harmful decrease of microbial diversity. If necessary, countermeasures are required, to maintain a healthy, diverse balance of beneficial, neutral and opportunistic pathogenic microorganisms. Our results serve as an important data collection for (i) future risk estimations of crewed space flight, (ii) an optimized design and planning of a spacecraft mission and (iii) for the selection of appropriate microbial monitoring approaches and potential countermeasures, to ensure a microbiologically safe space-flight environment.
Keywords: Built environment; Mars flight simulation; Mars500; Microbiota.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
References
-
- Gagarin YA, Lebedev VI. Survival in space. New York: F.A. Praeger; 1969.
-
- Manned Exploration Requirements and Considerations . Advanced Studies Office, Engineering and Development Directorate. Houston, Texas: NASA Manned Spacecraft Center; 1971. pp. 1–8.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

