Mechanism of mRNA-STAR domain interaction: Molecular dynamics simulations of Mammalian Quaking STAR protein
- PMID: 28974714
- PMCID: PMC5626755
- DOI: 10.1038/s41598-017-12930-2
Mechanism of mRNA-STAR domain interaction: Molecular dynamics simulations of Mammalian Quaking STAR protein
Abstract
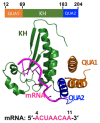
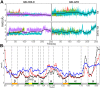
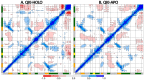
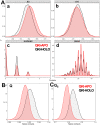
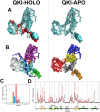
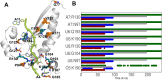
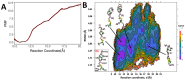
STAR proteins are evolutionary conserved mRNA-binding proteins that post-transcriptionally regulate gene expression at all stages of RNA metabolism. These proteins possess conserved STAR domain that recognizes identical RNA regulatory elements as YUAAY. Recently reported crystal structures show that STAR domain is composed of N-terminal QUA1, K-homology domain (KH) and C-terminal QUA2, and mRNA binding is mediated by KH-QUA2 domain. Here, we present simulation studies done to investigate binding of mRNA to STAR protein, mammalian Quaking protein (QKI). We carried out conventional MD simulations of STAR domain in presence and absence of mRNA, and studied the impact of mRNA on the stability, dynamics and underlying allosteric mechanism of STAR domain. Our unbiased simulations results show that presence of mRNA stabilizes the overall STAR domain by reducing the structural deviations, correlating the 'within-domain' motions, and maintaining the native contacts information. Absence of mRNA not only influenced the essential modes of motion of STAR domain, but also affected the connectivity of networks within STAR domain. We further explored the dissociation of mRNA from STAR domain using umbrella sampling simulations, and the results suggest that mRNA binding to STAR domain occurs in multi-step: first conformational selection of mRNA backbone conformations, followed by induced fit mechanism as nucleobases interact with STAR domain.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

