Targeting the alternative sigma factor RpoN to combat virulence in Pseudomonas aeruginosa
- PMID: 28974743
- PMCID: PMC5626770
- DOI: 10.1038/s41598-017-12667-y
Targeting the alternative sigma factor RpoN to combat virulence in Pseudomonas aeruginosa
Abstract
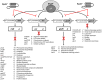
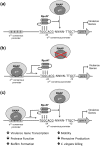
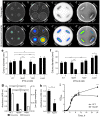
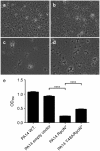
Pseudomonas aeruginosa is a Gram-negative, opportunistic pathogen that infects immunocompromised and cystic fibrosis patients. Treatment is difficult due to antibiotic resistance, and new antimicrobials are needed to treat infections. The alternative sigma factor 54 (σ54, RpoN), regulates many virulence-associated genes. Thus, we evaluated inhibition of virulence in P. aeruginosa by a designed peptide (RpoN molecular roadblock, RpoN*) which binds specifically to RpoN consensus promoters. We expected that RpoN* binding to its consensus promoter sites would repress gene expression and thus virulence by blocking RpoN and/or other transcription factors. RpoN* reduced transcription of approximately 700 genes as determined by microarray analysis, including genes related to virulence. RpoN* expression significantly reduced motility, protease secretion, pyocyanin and pyoverdine production, rhamnolipid production, and biofilm formation. Given the effectiveness of RpoN* in vitro, we explored its effects in a Caenorhabditis elegans-P. aeruginosa infection model. Expression of RpoN* protected C. elegans in a paralytic killing assay, whereas worms succumbed to paralysis and death in its absence. In a slow killing assay, which mimics establishment and proliferation of an infection, C. elegans survival was prolonged when RpoN* was expressed. Thus, blocking RpoN consensus promoter sites is an effective strategy for abrogation of P. aeruginosa virulence.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

Similar articles
-
Blocking RpoN reduces virulence of Pseudomonas aeruginosa isolated from cystic fibrosis patients and increases antibiotic sensitivity in a laboratory strain.Sci Rep. 2019 Apr 30;9(1):6677. doi: 10.1038/s41598-019-43060-6. Sci Rep. 2019. PMID: 31040330 Free PMC article.
-
RpoN-Dependent Direct Regulation of Quorum Sensing and the Type VI Secretion System in Pseudomonas aeruginosa PAO1.J Bacteriol. 2018 Jul 25;200(16):e00205-18. doi: 10.1128/JB.00205-18. Print 2018 Aug 15. J Bacteriol. 2018. PMID: 29760208 Free PMC article.
-
Analysis of the Pseudomonas aeruginosa regulon controlled by the sensor kinase KinB and sigma factor RpoN.J Bacteriol. 2012 Mar;194(6):1317-30. doi: 10.1128/JB.06105-11. Epub 2011 Dec 30. J Bacteriol. 2012. PMID: 22210761 Free PMC article.
-
Caenorhabditis elegans: a model genetic host to study Pseudomonas aeruginosa pathogenesis.Curr Opin Microbiol. 2000 Feb;3(1):29-34. doi: 10.1016/s1369-5274(99)00047-8. Curr Opin Microbiol. 2000. PMID: 10679415 Review.
-
Conversion of Pseudomonas aeruginosa to mucoidy in cystic fibrosis: environmental stress and regulation of bacterial virulence by alternative sigma factors.J Bacteriol. 1994 May;176(10):2773-80. doi: 10.1128/jb.176.10.2773-2780.1994. J Bacteriol. 1994. PMID: 8188579 Free PMC article. Review. No abstract available.
Cited by
-
Cross-resistance is modular in bacteria-phage interactions.PLoS Biol. 2018 Oct 3;16(10):e2006057. doi: 10.1371/journal.pbio.2006057. eCollection 2018 Oct. PLoS Biol. 2018. PMID: 30281587 Free PMC article.
-
Blocking RpoN reduces virulence of Pseudomonas aeruginosa isolated from cystic fibrosis patients and increases antibiotic sensitivity in a laboratory strain.Sci Rep. 2019 Apr 30;9(1):6677. doi: 10.1038/s41598-019-43060-6. Sci Rep. 2019. PMID: 31040330 Free PMC article.
-
Involvement of RpoN in Regulating Motility, Biofilm, Resistance, and Spoilage Potential of Pseudomonas fluorescens.Front Microbiol. 2021 May 31;12:641844. doi: 10.3389/fmicb.2021.641844. eCollection 2021. Front Microbiol. 2021. PMID: 34135871 Free PMC article.
-
Genome-Scale Mapping Reveals Complex Regulatory Activities of RpoN in Yersinia pseudotuberculosis.mSystems. 2020 Nov 10;5(6):e01006-20. doi: 10.1128/mSystems.01006-20. mSystems. 2020. PMID: 33172972 Free PMC article.
-
The sigma factor σ54 (rpoN) functions as a global regulator of antibiotic resistance, motility, metabolism, and virulence in Clostridioides difficile.Front Microbiol. 2025 Apr 29;16:1569627. doi: 10.3389/fmicb.2025.1569627. eCollection 2025. Front Microbiol. 2025. PMID: 40365067 Free PMC article.
References
-
- Registry, C. F. F. P. 2015 Annual Data Report. (2016).
-
- Porras-Gomez M, Vega-Baudrit J, Nunez-Corrales S. Overview of Multidrug-Resistant Pseudomonas aeruginosa and Novel Therapeutic Approaches. Journal of Biomaterials and Nanobiotechnology. 2012;3:9. doi: 10.4236/jbnb.2012.324053. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

