Neofunctionalization of "Juvenile Hormone Esterase Duplication" in Drosophila as an odorant-degrading enzyme towards food odorants
- PMID: 28974761
- PMCID: PMC5626784
- DOI: 10.1038/s41598-017-13015-w
Neofunctionalization of "Juvenile Hormone Esterase Duplication" in Drosophila as an odorant-degrading enzyme towards food odorants
Abstract
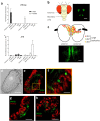
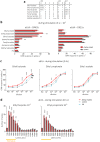
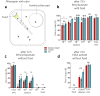
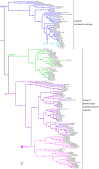
Odorant degrading enzymes (ODEs) are thought to be responsible, at least in part, for olfactory signal termination in the chemosensory system by rapid degradation of odorants in the vicinity of the receptors. A carboxylesterase, specifically expressed in Drosophila antennae, called "juvenile hormone esterase duplication (JHEdup)" has been previously reported to hydrolyse different fruit esters in vitro. Here we functionally characterize JHEdup in vivo. We show that the jhedup gene is highly expressed in large basiconic sensilla that have been reported to detect several food esters. An electrophysiological analysis demonstrates that ab1A olfactory neurons of jhedup mutant flies exhibit an increased response to certain food acetates. Furthermore, mutant flies show a higher sensitivity towards the same odorants in behavioural assays. A phylogenetic analysis reveals that jhedup arose as a duplication of the juvenile hormone esterase gene during the evolution of Diptera, most likely in the ancestor of Schizophora, and has been conserved in all the 12 sequenced Drosophila species. Jhedup exhibits also an olfactory-predominant expression pattern in other Drosophila species. Our results support the implication of JHEdup in the degradation of food odorants in D. melanogaster and propose a neofunctionalization of this enzyme as a bona fide ODE in Drosophilids.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



Similar articles
-
A carboxylesterase, Esterase-6, modulates sensory physiological and behavioral response dynamics to pheromone in Drosophila.BMC Biol. 2012 Jun 21;10:56. doi: 10.1186/1741-7007-10-56. BMC Biol. 2012. PMID: 22715942 Free PMC article.
-
Identification of candidate odorant degrading gene/enzyme systems in the antennal transcriptome of Drosophila melanogaster.Insect Biochem Mol Biol. 2014 Oct;53:30-43. doi: 10.1016/j.ibmb.2014.07.003. Epub 2014 Jul 16. Insect Biochem Mol Biol. 2014. PMID: 25038463
-
The molecular basis for the neofunctionalization of the juvenile hormone esterase duplication in Drosophila.Insect Biochem Mol Biol. 2019 Mar;106:10-18. doi: 10.1016/j.ibmb.2019.01.001. Epub 2019 Jan 3. Insect Biochem Mol Biol. 2019. PMID: 30611903
-
Elements of olfactory reception in adult Drosophila melanogaster.Anat Rec (Hoboken). 2013 Sep;296(9):1477-88. doi: 10.1002/ar.22747. Epub 2013 Jul 31. Anat Rec (Hoboken). 2013. PMID: 23904114 Review.
-
Evolving olfactory systems on the fly.Trends Genet. 2010 Jul;26(7):307-16. doi: 10.1016/j.tig.2010.04.004. Epub 2010 May 27. Trends Genet. 2010. PMID: 20537755 Review.
Cited by
-
Pheromone sensing in Drosophila requires support cell-expressed Osiris 8.BMC Biol. 2022 Oct 11;20(1):230. doi: 10.1186/s12915-022-01425-w. BMC Biol. 2022. PMID: 36217142 Free PMC article.
-
Antennal transcriptome analysis reveals sensory receptors potentially associated with host detection in the livestock pest Lucilia cuprina.Parasit Vectors. 2024 Jul 18;17(1):308. doi: 10.1186/s13071-024-06391-6. Parasit Vectors. 2024. PMID: 39026238 Free PMC article.
-
Molecular Profiling of the Drosophila Antenna Reveals Conserved Genes Underlying Olfaction in Insects.G3 (Bethesda). 2019 Nov 5;9(11):3753-3771. doi: 10.1534/g3.119.400669. G3 (Bethesda). 2019. PMID: 31527046 Free PMC article.
-
Antennal transcriptomic analysis of carboxylesterases and glutathione S-transferases associated with odorant degradation in the tea gray geometrid, Ectropis grisescens (Lepidoptera, Geometridae).Front Physiol. 2023 Apr 4;14:1183610. doi: 10.3389/fphys.2023.1183610. eCollection 2023. Front Physiol. 2023. PMID: 37082242 Free PMC article.
-
Transcriptome Analysis and Knockdown of the Juvenile Hormone Esterase Gene Reveal Abnormal Feeding Behavior in the Sugarcane Giant Borer.Front Physiol. 2020 Oct 28;11:588450. doi: 10.3389/fphys.2020.588450. eCollection 2020. Front Physiol. 2020. PMID: 33192604 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

