The free-energy cost of interaction between DNA loops
- PMID: 28974770
- PMCID: PMC5626758
- DOI: 10.1038/s41598-017-12765-x
The free-energy cost of interaction between DNA loops
Abstract
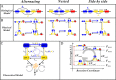
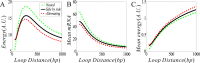
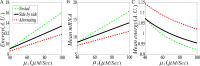
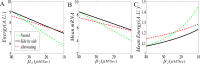
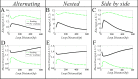
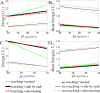
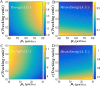
From the viewpoint of thermodynamics, the formation of DNA loops and the interaction between them, which are all non-equilibrium processes, result in the change of free energy, affecting gene expression and further cell-to-cell variability as observed experimentally. However, how these processes dissipate free energy remains largely unclear. Here, by analyzing a mechanic model that maps three fundamental topologies of two interacting DNA loops into a 4-state model of gene transcription, we first show that a longer DNA loop needs more mean free energy consumption. Then, independent of the type of interacting two DNA loops (nested, side-by-side or alternating), the promotion between them always consumes less mean free energy whereas the suppression dissipates more mean free energy. More interestingly, we find that in contrast to the mechanism of direct looping between promoter and enhancer, the facilitated-tracking mechanism dissipates less mean free energy but enhances the mean mRNA expression, justifying the facilitated-tracking hypothesis, a long-standing debate in biology. Based on minimal energy principle, we thus speculate that organisms would utilize the mechanisms of loop-loop promotion and facilitated tracking to survive in complex environments. Our studies provide insights into the understanding of gene expression regulation mechanism from the view of energy consumption.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

