Genome-wide identification and expression analyses of the homeobox transcription factor family during ovule development in seedless and seeded grapes
- PMID: 28974771
- PMCID: PMC5626728
- DOI: 10.1038/s41598-017-12988-y
Genome-wide identification and expression analyses of the homeobox transcription factor family during ovule development in seedless and seeded grapes
Erratum in
-
Publisher Correction: Genome-wide identification and expression analyses of the homeobox transcription factor family during ovule development in seedless and seeded grapes.Sci Rep. 2017 Nov 17;7(1):16067. doi: 10.1038/s41598-017-16207-6. Sci Rep. 2017. PMID: 29150663 Free PMC article.
Abstract
Seedless grapes are of considerable importance for the raisin and table grape industries. Previous transcriptome analyses of seed development in grape revealed that genes encoding homeobox transcription factors were differentially regulated in seedless compared with seeded grape during seed development. In the present study, we identified a total of 73 homeobox-like genes in the grapevine genome and analyzed the genomic content and expression profiles of these genes. Based on domain architecture and phylogenetic analyses grape homeobox genes can be classified into eleven subfamilies. An analysis of the exon-intron structures and conserved motifs provided further insight into the evolutionary relationships between these genes. Evaluation of synteny indicated that segmental and tandem duplications have contributed greatly to the expansion of the grape homeobox gene superfamily. Synteny analysis between the grape and Arabidopsis genomes provided a potential functional relevance for these genes. The tissue-specific expression patterns of homeobox genes suggested roles in both vegetative and reproductive tissues. Expression profiling of these genes during the course of ovule development in seeded and seedless cultivars suggested a potential role in ovule abortion associated with seedlessness. This study will facilitate the functional analysis of these genes and provide new resources for molecular breeding of seedless grapes.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
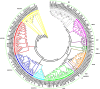
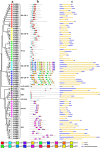
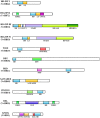
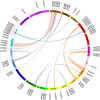
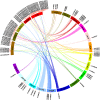
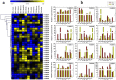
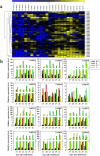
Similar articles
-
Evolutionary and expression analysis of a MADS-box gene superfamily involved in ovule development of seeded and seedless grapevines.Mol Genet Genomics. 2015 Jun;290(3):825-46. doi: 10.1007/s00438-014-0961-y. Epub 2014 Nov 28. Mol Genet Genomics. 2015. PMID: 25429734
-
Genome-wide identification and expression analysis reveal the potential function of ethylene responsive factor gene family in response to Botrytis cinerea infection and ovule development in grapes (Vitis vinifera L.).Plant Biol (Stuttg). 2019 Jul;21(4):571-584. doi: 10.1111/plb.12943. Epub 2018 Dec 13. Plant Biol (Stuttg). 2019. PMID: 30468551
-
Genome-Wide Characterization and Expression Profiling of GASA Genes during Different Stages of Seed Development in Grapevine (Vitis vinifera L.) Predict Their Involvement in Seed Development.Int J Mol Sci. 2020 Feb 6;21(3):1088. doi: 10.3390/ijms21031088. Int J Mol Sci. 2020. PMID: 32041336 Free PMC article.
-
Seedlessness Trait and Genome Editing-A Review.Int J Mol Sci. 2023 Mar 16;24(6):5660. doi: 10.3390/ijms24065660. Int J Mol Sci. 2023. PMID: 36982733 Free PMC article. Review.
-
Establishing a framework for female germline initiation in the plant ovule.J Exp Bot. 2019 Jun 1;70(11):2937-2949. doi: 10.1093/jxb/erz212. J Exp Bot. 2019. PMID: 31063548 Review.
Cited by
-
Identification and Characterization of the WOX Family Genes in Five Solanaceae Species Reveal Their Conserved Roles in Peptide Signaling.Genes (Basel). 2018 May 17;9(5):260. doi: 10.3390/genes9050260. Genes (Basel). 2018. PMID: 29772825 Free PMC article.
-
Genome-Wide Identification and Characterization of TCP Gene Family Members in Melastoma candidum.Molecules. 2022 Dec 18;27(24):9036. doi: 10.3390/molecules27249036. Molecules. 2022. PMID: 36558169 Free PMC article.
-
Genome-Wide Identification and Expression Analysis of the Basic Leucine Zipper (bZIP) Transcription Factor Gene Family in Fusarium graminearum.Genes (Basel). 2022 Mar 28;13(4):607. doi: 10.3390/genes13040607. Genes (Basel). 2022. PMID: 35456413 Free PMC article.
-
Identification of Homeobox Genes Associated with Lignification and Their Expression Patterns in Bamboo Shoots.Biomolecules. 2019 Dec 11;9(12):862. doi: 10.3390/biom9120862. Biomolecules. 2019. PMID: 31835882 Free PMC article.
-
Genome-Wide Analysis of the Homeobox Gene Family and Identification of Drought-Responsive Members in Populus trichocarpa.Plants (Basel). 2021 Oct 25;10(11):2284. doi: 10.3390/plants10112284. Plants (Basel). 2021. PMID: 34834651 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

