A mutation in an exoglucanase of Xanthomonas oryzae pv. oryzae, which confers an endo mode of activity, affects bacterial virulence, but not the induction of immune responses, in rice
- PMID: 28976110
- PMCID: PMC6638110
- DOI: 10.1111/mpp.12620
A mutation in an exoglucanase of Xanthomonas oryzae pv. oryzae, which confers an endo mode of activity, affects bacterial virulence, but not the induction of immune responses, in rice
Abstract
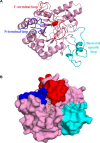
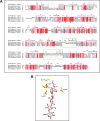
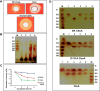
Xanthomonas oryzae pv. oryzae (Xoo) causes bacterial blight, a serious disease of rice. Xoo secretes a repertoire of cell wall-degrading enzymes, including cellulases, xylanases and pectinases, to degrade various polysaccharide components of the rice cell wall. A secreted Xoo cellulase, CbsA, is not only a key virulence factor of Xoo, but is also a potent inducer of innate immune responses of rice. In this study, we solved the crystal structure of the catalytic domain of the CbsA protein to a resolution of 1.86 Å. The core structure of CbsA shows a central distorted TIM barrel made up of eight β strands with N- and C-terminal loops enclosing the active site, which is a characteristic structural feature of an exoglucanase. The aspartic acid at the 131st position of CbsA was predicted to be important for catalysis and was therefore mutated to alanine to study its role in the catalysis and biological functions of CbsA. Intriguingly, the D131A CbsA mutant protein displayed the enzymatic activity of a typical endoglucanase. D131A CbsA was as proficient as wild-type (Wt) CbsA in inducing rice immune responses, but was deficient in virulence-promoting activity. This indicates that the specific exoglucanase activity of the Wt CbsA protein is required for this protein to promote the growth of Xoo in rice.
Keywords: Xanthomonas oryzae pv. oryzae; cellulases; endoglucanase; exoglucanase; rice.
© 2017 BSPP AND JOHN WILEY & SONS LTD.
Figures

Similar articles
-
Role of the FnIII domain associated with a cell wall-degrading enzyme cellobiosidase of Xanthomonas oryzae pv. oryzae.Mol Plant Pathol. 2022 Jul;23(7):1011-1021. doi: 10.1111/mpp.13205. Epub 2022 Mar 12. Mol Plant Pathol. 2022. PMID: 35278018 Free PMC article.
-
Mutational analysis of predicted active site residues of an exoglucanase secreted by Xanthomonas oryzae pv. oryzae to determine their role in catalysis and in virulence on rice.Enzyme Microb Technol. 2024 Mar;174:110372. doi: 10.1016/j.enzmictec.2023.110372. Epub 2023 Dec 6. Enzyme Microb Technol. 2024. PMID: 38104475
-
Action of Multiple Cell Wall-Degrading Enzymes Is Required for Elicitation of Innate Immune Responses During Xanthomonas oryzae pv. oryzae Infection in Rice.Mol Plant Microbe Interact. 2016 Aug;29(8):599-608. doi: 10.1094/MPMI-02-16-0039-R. Epub 2016 Jul 19. Mol Plant Microbe Interact. 2016. PMID: 27269510
-
Molecular determinants of disease and resistance in interactions of Xanthomonas oryzae pv. oryzae and rice.Microbes Infect. 2002 Nov;4(13):1361-7. doi: 10.1016/s1286-4579(02)00004-7. Microbes Infect. 2002. PMID: 12443901 Review.
-
A review of approaches to control bacterial leaf blight in rice.World J Microbiol Biotechnol. 2022 May 17;38(7):113. doi: 10.1007/s11274-022-03298-1. World J Microbiol Biotechnol. 2022. PMID: 35578069 Review.
Cited by
-
Transcriptional regulator Sar regulates the multiple secretion systems in Xanthomonas oryzae.Mol Plant Pathol. 2023 Jan;24(1):16-27. doi: 10.1111/mpp.13272. Epub 2022 Sep 30. Mol Plant Pathol. 2023. PMID: 36177860 Free PMC article.
-
Role of the FnIII domain associated with a cell wall-degrading enzyme cellobiosidase of Xanthomonas oryzae pv. oryzae.Mol Plant Pathol. 2022 Jul;23(7):1011-1021. doi: 10.1111/mpp.13205. Epub 2022 Mar 12. Mol Plant Pathol. 2022. PMID: 35278018 Free PMC article.
-
Xanthomonas oryzae pv. oryzae XopQ protein suppresses rice immune responses through interaction with two 14-3-3 proteins but its phospho-null mutant induces rice immune responses and interacts with another 14-3-3 protein.Mol Plant Pathol. 2019 Jul;20(7):976-989. doi: 10.1111/mpp.12807. Epub 2019 May 15. Mol Plant Pathol. 2019. PMID: 31094082 Free PMC article.
-
An Emerging Bacterial Leaf Disease in Rice Caused by Pantoea ananatis and Pantoea eucalypti in Northeast China.Microorganisms. 2025 Jun 13;13(6):1376. doi: 10.3390/microorganisms13061376. Microorganisms. 2025. PMID: 40572264 Free PMC article.
-
Arms and ammunitions: effectors at the interface of rice and it's pathogens and pests.Rice (N Y). 2021 Nov 18;14(1):94. doi: 10.1186/s12284-021-00534-4. Rice (N Y). 2021. PMID: 34792681 Free PMC article. Review.
References
-
- Adams, P.D. , Afonine, P.V. , Bunkóczi, G. , Chen, V.B. , Davis, I.W. , Echols, N. , Headd, J.J. , Hung, L.‐W. , Kapral, G.J. , Grosse‐Kunstleve, R.W. , McCoy, A.J. , Moriarty, N.W. , Oeffner, R. , Read, R.J. , Richardson, D.C. , Richardson, J.S. , Terwilliger, T.C. and Zwart, P.H. (2010) PHENIX: a comprehensive Python‐based system for macromolecular structure solution. Acta Crystallogr. D: Biol. Crystallogr. 66, 213–221. - PMC - PubMed
-
- Collaborative Computational Project . (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D: Biol. Crystallogr. 50, 760. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

