Cancer drug addiction is relayed by an ERK2-dependent phenotype switch
- PMID: 28976960
- PMCID: PMC5640985
- DOI: 10.1038/nature24037
Cancer drug addiction is relayed by an ERK2-dependent phenotype switch
Abstract
Observations from cultured cells, animal models and patients raise the possibility that the dependency of tumours on the therapeutic drugs to which they have acquired resistance represents a vulnerability with potential applications in cancer treatment. However, for this drug addiction trait to become of clinical interest, we must first define the mechanism that underlies it. We performed an unbiased CRISPR-Cas9 knockout screen on melanoma cells that were both resistant and addicted to inhibition of the serine/threonine-protein kinase BRAF, in order to functionally mine their genome for 'addiction genes'. Here we describe a signalling pathway comprising ERK2 kinase and JUNB and FRA1 transcription factors, disruption of which allowed addicted tumour cells to survive on treatment discontinuation. This occurred in both cultured cells and mice and was irrespective of the acquired drug resistance mechanism. In melanoma and lung cancer cells, death induced by drug withdrawal was preceded by a specific ERK2-dependent phenotype switch, alongside transcriptional reprogramming reminiscent of the epithelial-mesenchymal transition. In melanoma cells, this reprogramming caused the shutdown of microphthalmia-associated transcription factor (MITF), a lineage survival oncoprotein; restoring this protein reversed phenotype switching and prevented the lethality associated with drug addiction. In patients with melanoma that had progressed during treatment with a BRAF inhibitor, treatment cessation was followed by increased expression of the receptor tyrosine kinase AXL, which is associated with the phenotype switch. Drug discontinuation synergized with the melanoma chemotherapeutic agent dacarbazine by further suppressing MITF and its prosurvival target, B-cell lymphoma 2 (BCL-2), and by inducing DNA damage in cancer cells. Our results uncover a pathway that underpins drug addiction in cancer cells, which may help to guide the use of alternating therapeutic strategies for enhanced clinical responses in drug-resistant cancers.
Conflict of interest statement
The authors declare no competing financial interests.
Figures






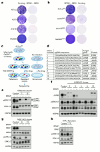
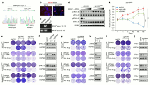
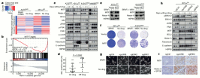
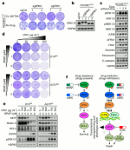
Comment in
-
Cancer: Tumours addicted to drugs are vulnerable.Nature. 2017 Oct 12;550(7675):192-193. doi: 10.1038/nature24148. Epub 2017 Oct 4. Nature. 2017. PMID: 28976969 No abstract available.
-
Targeted therapies: Understanding tumour drug addiction.Nat Rev Cancer. 2017 Oct 25;17(11):634-635. doi: 10.1038/nrc.2017.98. Nat Rev Cancer. 2017. PMID: 29068004 No abstract available.
References
-
- Suda K, et al. Conversion from the "oncogene addiction" to ‘drug addiction’ by intensive inhibition of the EGFR and MET in lung cancer with activating EGFR mutation. Lung Cancer. 2012;76:292–299. - PubMed
-
- Sun C, et al. Reversible and adaptive resistance to BRAF(V600E) inhibition in melanoma. Nature. 2014;508:118–122. - PubMed
-
- Seifert H, et al. Prognostic markers and tumour growth kinetics in melanoma patients progressing on vemurafenib. Melanoma Res. 2016;26:138–144. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

