Cytoplasmic chromatin triggers inflammation in senescence and cancer
- PMID: 28976970
- PMCID: PMC5850938
- DOI: 10.1038/nature24050
Cytoplasmic chromatin triggers inflammation in senescence and cancer
Abstract
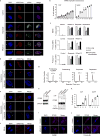
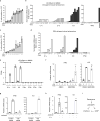
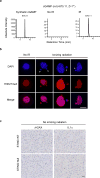
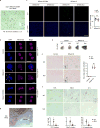
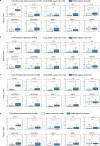
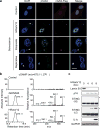
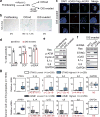
Chromatin is traditionally viewed as a nuclear entity that regulates gene expression and silencing. However, we recently discovered the presence of cytoplasmic chromatin fragments that pinch off from intact nuclei of primary cells during senescence, a form of terminal cell-cycle arrest associated with pro-inflammatory responses. The functional significance of chromatin in the cytoplasm is unclear. Here we show that cytoplasmic chromatin activates the innate immunity cytosolic DNA-sensing cGAS-STING (cyclic GMP-AMP synthase linked to stimulator of interferon genes) pathway, leading both to short-term inflammation to restrain activated oncogenes and to chronic inflammation that associates with tissue destruction and cancer. The cytoplasmic chromatin-cGAS-STING pathway promotes the senescence-associated secretory phenotype in primary human cells and in mice. Mice deficient in STING show impaired immuno-surveillance of oncogenic RAS and reduced tissue inflammation upon ionizing radiation. Furthermore, this pathway is activated in cancer cells, and correlates with pro-inflammatory gene expression in human cancers. Overall, our findings indicate that genomic DNA serves as a reservoir to initiate a pro-inflammatory pathway in the cytoplasm in senescence and cancer. Targeting the cytoplasmic chromatin-mediated pathway may hold promise in treating inflammation-related disorders.
Figures







Comment in
-
Cancer biology: Genome jail-break triggers lockdown.Nature. 2017 Oct 19;550(7676):340-341. doi: 10.1038/nature24146. Epub 2017 Oct 4. Nature. 2017. PMID: 28976957 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

