Transcriptome analysis of genes involved in defense against alkaline stress in roots of wild jujube (Ziziphus acidojujuba)
- PMID: 28976994
- PMCID: PMC5627934
- DOI: 10.1371/journal.pone.0185732
Transcriptome analysis of genes involved in defense against alkaline stress in roots of wild jujube (Ziziphus acidojujuba)
Abstract
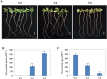
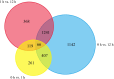
Wild jujube (Ziziphus acidojujuba Mill.) is highly tolerant to alkaline, saline and drought stress; however, no studies have performed transcriptome profiling to study the response of wild jujube to these and other abiotic stresses. In this study, we examined the tolerance of wild jujube to NaHCO3-NaOH solution and analyzed gene expression profiles in response to alkaline stress. Physiological experiments revealed that H2O2 content in leaves increased significantly and root activity decreased quickly during alkaline of pH 9.5 treatment. For transcriptome analysis, wild jujube plants grown hydroponically were treated with NaHCO3-NaOH solution for 0, 1, and 12 h and six transcriptomes from roots were built. In total, 32,758 genes were generated, and 3,604 differentially expressed genes (DEGs) were identified. After 1 h, 853 genes showed significantly different expression between control and treated plants; after 12 h, expression of 2,856 genes was significantly different. The expression pattern of nine genes was validated by quantitative real-time PCR. After gene annotation and gene ontology enrichment analysis, the genes encoding transcriptional factors, serine/threonine-protein kinases, heat shock proteins, cysteine-like kinases, calmodulin-like proteins, and reactive oxygen species (ROS) scavengers were found to be closely involved in alkaline stress response. These results will provide useful insights for elucidating the mechanisms underlying alkaline tolerance in wild jujube.
Conflict of interest statement
Figures



Similar articles
-
Comparative transcriptome analysis uncovers different heat stress responses in heat-resistant and heat-sensitive jujube cultivars.PLoS One. 2020 Sep 21;15(9):e0235763. doi: 10.1371/journal.pone.0235763. eCollection 2020. PLoS One. 2020. PMID: 32956359 Free PMC article.
-
Transcriptome profiling analysis revealed co-regulation of multiple pathways in jujube during infection by 'Candidatus Phytoplasma ziziphi'.Gene. 2018 Jul 30;665:82-95. doi: 10.1016/j.gene.2018.04.070. Epub 2018 Apr 27. Gene. 2018. PMID: 29709641
-
Genome-Wide Identification of WRKY Transcription Factors in Chinese jujube (Ziziphus jujuba Mill.) and Their Involvement in Fruit Developing, Ripening, and Abiotic Stress.Genes (Basel). 2019 May 10;10(5):360. doi: 10.3390/genes10050360. Genes (Basel). 2019. PMID: 31083435 Free PMC article.
-
Genome-wide identification, characterization, and expression analysis of the expansin gene family in Chinese jujube (Ziziphus jujuba Mill.).Planta. 2019 Mar;249(3):815-829. doi: 10.1007/s00425-018-3020-9. Epub 2018 Nov 8. Planta. 2019. PMID: 30411169
-
Triterpenoids in Jujube: A Review of Composition, Content Diversity, Pharmacological Effects, Synthetic Pathway, and Variation during Domestication.Plants (Basel). 2023 Mar 29;12(7):1501. doi: 10.3390/plants12071501. Plants (Basel). 2023. PMID: 37050126 Free PMC article. Review.
Cited by
-
Comparative transcriptome analysis reveals gene expression differences between two peach cultivars under saline-alkaline stress.Hereditas. 2020 Mar 31;157(1):9. doi: 10.1186/s41065-020-00122-4. Hereditas. 2020. PMID: 32234076 Free PMC article.
-
Transcriptome Analysis of Jujube (Ziziphus jujuba Mill.) Response to Heat Stress.Int J Genomics. 2021 Dec 2;2021:3442277. doi: 10.1155/2021/3442277. eCollection 2021. Int J Genomics. 2021. PMID: 34901262 Free PMC article.
-
Physiological and transcriptomic analyses of yellow horn (Xanthoceras sorbifolia) provide important insights into salt and saline-alkali stress tolerance.PLoS One. 2020 Dec 22;15(12):e0244365. doi: 10.1371/journal.pone.0244365. eCollection 2020. PLoS One. 2020. PMID: 33351842 Free PMC article.
-
Alkaline Stress Induces Different Physiological, Hormonal and Gene Expression Responses in Diploid and Autotetraploid Rice.Int J Mol Sci. 2022 May 16;23(10):5561. doi: 10.3390/ijms23105561. Int J Mol Sci. 2022. PMID: 35628377 Free PMC article.
-
Genome-wide analysis of the bZIP gene family in Chinese jujube (Ziziphus jujuba Mill.).BMC Genomics. 2020 Jul 14;21(1):483. doi: 10.1186/s12864-020-06890-7. BMC Genomics. 2020. PMID: 32664853 Free PMC article.
References
-
- Zhang X, Takano T, Liu S. Identification of a mitochondrial ATP synthase small subunit gene (RMtATP6) expressed in response to salts and osmotic stresses in rice (Oryza sativa L.). J Exp Bot. 2006; 57: 193–200. doi: 10.1093/jxb/erj025 - DOI - PubMed
-
- Shi DC, Yin LJ. Difference between salt (NaCl) and alkaline (Na2CO3) stresses on Puccinellia tenuiflora (Griseb.) Scribn. et Merr. plants. Acta Bot Sin. 1993; 35: 144–149.
-
- Takahashi M, Nakanishi H, Kawasaki S, Nishizawa NK, Mori S. Enhanced tolerance of rice to low iron availability in alkaline soils using barley nicotianamine aminotransferase genes. Nat Biotechnol. 2001; 19: 466–469. doi: 10.1038/88143 - DOI - PubMed
-
- Ishimaru Y, Kim S, Tsukamoto T, Oki H, Kobayashi T, Watanabe S, et al. Mutational reconstructed ferric chelate reductase confers enhanced tolerance in rice to iron deficiency in calcareous soil. Proc Natl Acad Sci USA. 2007; 104: 7373–7378. doi: 10.1073/pnas.0610555104 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

